Tony Shen
Compositional Flows for 3D Molecule and Synthesis Pathway Co-design
Apr 10, 2025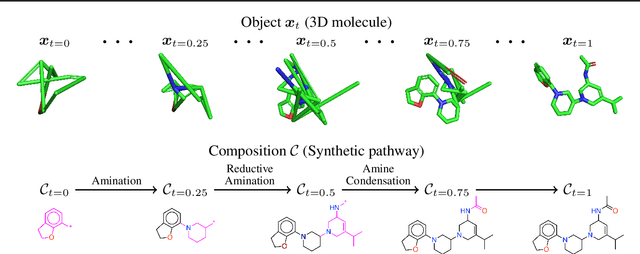
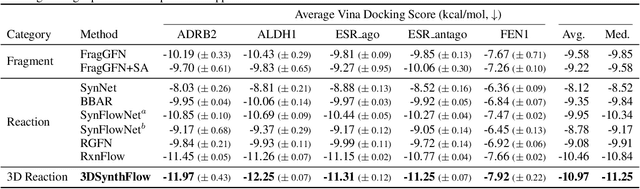
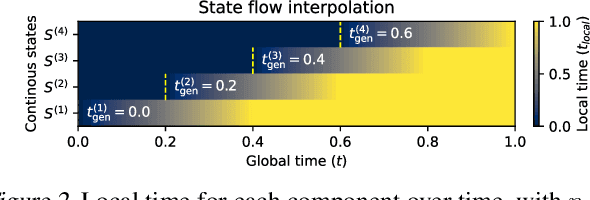
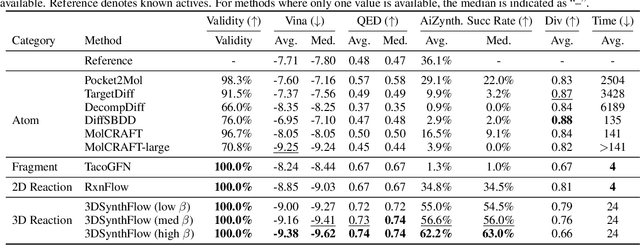
Abstract:Many generative applications, such as synthesis-based 3D molecular design, involve constructing compositional objects with continuous features. Here, we introduce Compositional Generative Flows (CGFlow), a novel framework that extends flow matching to generate objects in compositional steps while modeling continuous states. Our key insight is that modeling compositional state transitions can be formulated as a straightforward extension of the flow matching interpolation process. We further build upon the theoretical foundations of generative flow networks (GFlowNets), enabling reward-guided sampling of compositional structures. We apply CGFlow to synthesizable drug design by jointly designing the molecule's synthetic pathway with its 3D binding pose. Our approach achieves state-of-the-art binding affinity on all 15 targets from the LIT-PCBA benchmark, and 5.8$\times$ improvement in sampling efficiency compared to 2D synthesis-based baseline. To our best knowledge, our method is also the first to achieve state of-art-performance in both Vina Dock (-9.38) and AiZynth success rate (62.2\%) on the CrossDocked benchmark.
Generative Flows on Synthetic Pathway for Drug Design
Oct 06, 2024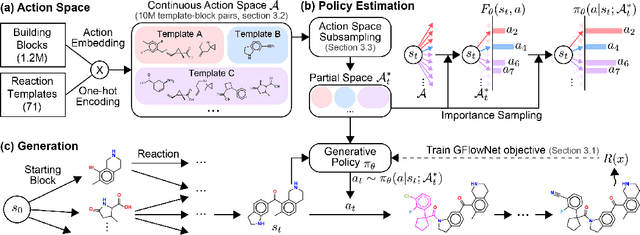
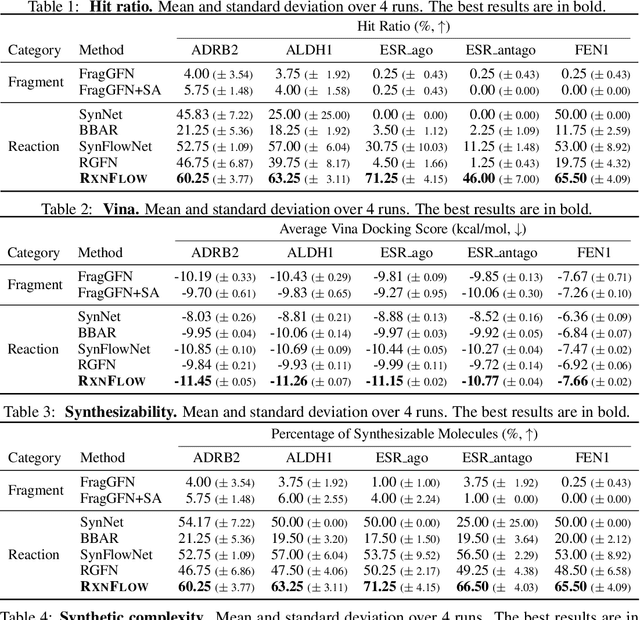
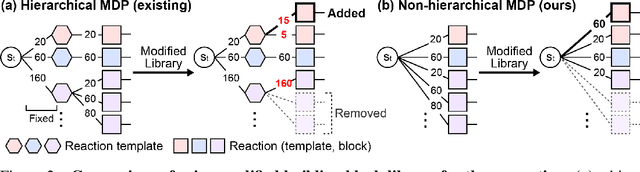
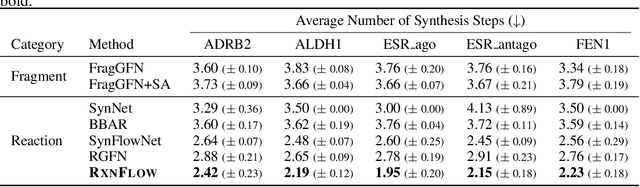
Abstract:Generative models in drug discovery have recently gained attention as efficient alternatives to brute-force virtual screening. However, most existing models do not account for synthesizability, limiting their practical use in real-world scenarios. In this paper, we propose RxnFlow, which sequentially assembles molecules using predefined molecular building blocks and chemical reaction templates to constrain the synthetic chemical pathway. We then train on this sequential generating process with the objective of generative flow networks (GFlowNets) to generate both highly rewarded and diverse molecules. To mitigate the large action space of synthetic pathways in GFlowNets, we implement a novel action space subsampling method. This enables RxnFlow to learn generative flows over extensive action spaces comprising combinations of 1.2 million building blocks and 71 reaction templates without significant computational overhead. Additionally, RxnFlow can employ modified or expanded action spaces for generation without retraining, allowing for the introduction of additional objectives or the incorporation of newly discovered building blocks. We experimentally demonstrate that RxnFlow outperforms existing reaction-based and fragment-based models in pocket-specific optimization across various target pockets. Furthermore, RxnFlow achieves state-of-the-art performance on CrossDocked2020 for pocket-conditional generation, with an average Vina score of -8.85kcal/mol and 34.8% synthesizability.
Geometric-informed GFlowNets for Structure-Based Drug Design
Jun 16, 2024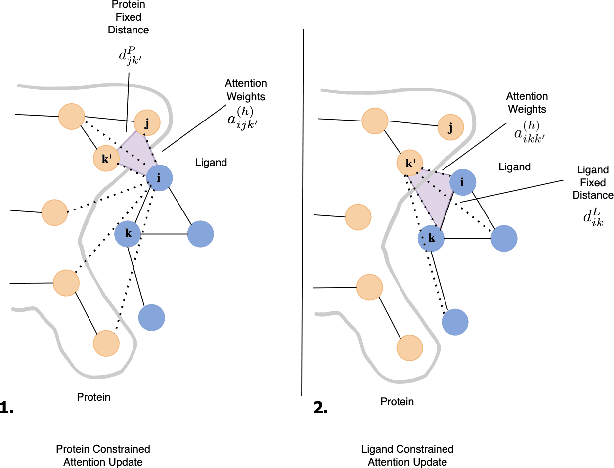


Abstract:The rise of cost involved with drug discovery and current speed of which they are discover, underscore the need for more efficient structure-based drug design (SBDD) methods. We employ Generative Flow Networks (GFlowNets), to effectively explore the vast combinatorial space of drug-like molecules, which traditional virtual screening methods fail to cover. We introduce a novel modification to the GFlowNet framework by incorporating trigonometrically consistent embeddings, previously utilized in tasks involving protein conformation and protein-ligand interactions, to enhance the model's ability to generate molecules tailored to specific protein pockets. We have modified the existing protein conditioning used by GFlowNets, blending geometric information from both protein and ligand embeddings to achieve more geometrically consistent embeddings. Experiments conducted using CrossDocked2020 demonstrated an improvement in the binding affinity between generated molecules and protein pockets for both single and multi-objective tasks, compared to previous work. Additionally, we propose future work aimed at further increasing the geometric information captured in protein-ligand interactions.
TacoGFN: Target Conditioned GFlowNet for Structure-Based Drug Design
Oct 05, 2023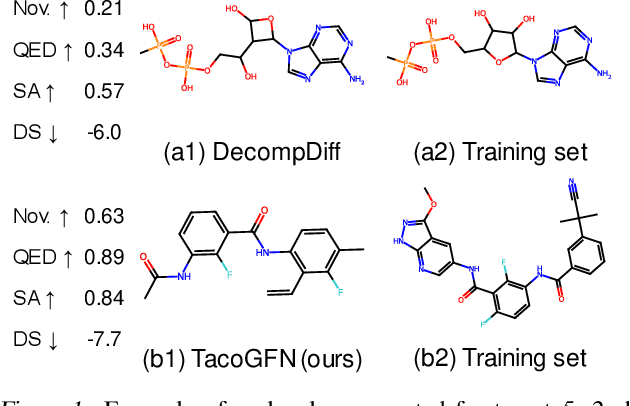



Abstract:We seek to automate the generation of drug-like compounds conditioned to specific protein pocket targets. Most current methods approximate the protein-molecule distribution of a finite dataset and, therefore struggle to generate molecules with significant binding improvement over the training dataset. We instead frame the pocket-conditioned molecular generation task as an RL problem and develop TacoGFN, a target conditional Generative Flow Network model. Our method is explicitly encouraged to generate molecules with desired properties as opposed to fitting on a pre-existing data distribution. To this end, we develop transformer-based docking score prediction to speed up docking score computation and propose TacoGFN to explore molecule space efficiently. Furthermore, we incorporate several rounds of active learning where generated samples are queried using a docking oracle to improve the docking score prediction. This approach allows us to accurately explore as much of the molecule landscape as we can afford computationally. Empirically, molecules generated using TacoGFN and its variants significantly outperform all baseline methods across every property (Docking score, QED, SA, Lipinski), while being orders of magnitude faster.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge