Mario Michael Krell
Extreme Acceleration of Graph Neural Network-based Prediction Models for Quantum Chemistry
Nov 25, 2022Abstract:Molecular property calculations are the bedrock of chemical physics. High-fidelity \textit{ab initio} modeling techniques for computing the molecular properties can be prohibitively expensive, and motivate the development of machine-learning models that make the same predictions more efficiently. Training graph neural networks over large molecular databases introduces unique computational challenges such as the need to process millions of small graphs with variable size and support communication patterns that are distinct from learning over large graphs such as social networks. This paper demonstrates a novel hardware-software co-design approach to scale up the training of graph neural networks for molecular property prediction. We introduce an algorithm to coalesce the batches of molecular graphs into fixed size packs to eliminate redundant computation and memory associated with alternative padding techniques and improve throughput via minimizing communication. We demonstrate the effectiveness of our co-design approach by providing an implementation of a well-established molecular property prediction model on the Graphcore Intelligence Processing Units (IPU). We evaluate the training performance on multiple molecular graph databases with varying degrees of graph counts, sizes and sparsity. We demonstrate that such a co-design approach can reduce the training time of such molecular property prediction models from days to less than two hours, opening new possibilities for AI-driven scientific discovery.
Tuple Packing: Efficient Batching of Small Graphs in Graph Neural Networks
Sep 18, 2022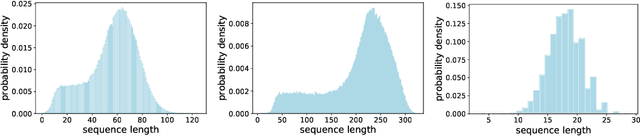
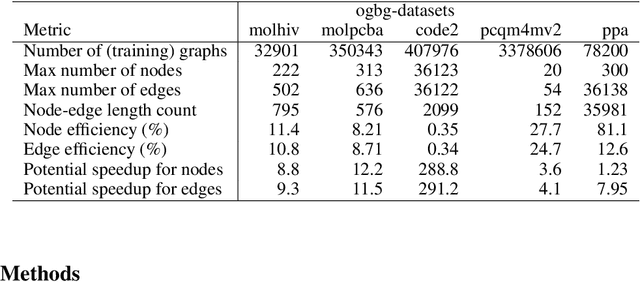
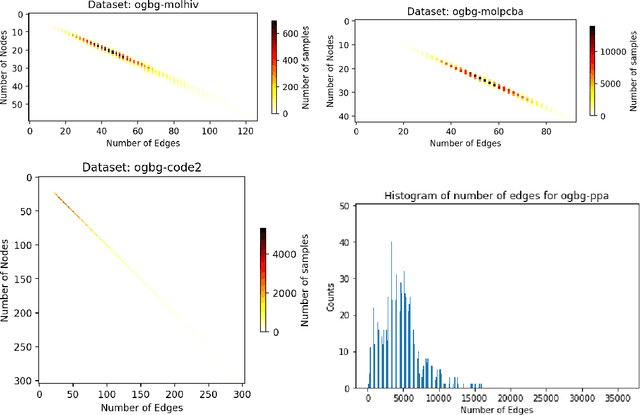
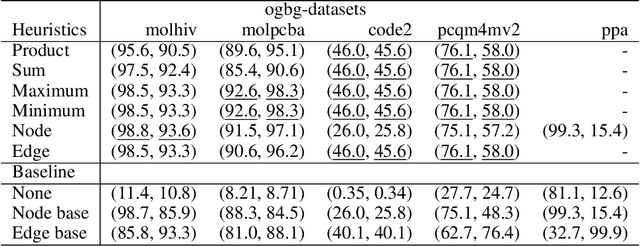
Abstract:When processing a batch of graphs in machine learning models such as Graph Neural Networks (GNN), it is common to combine several small graphs into one overall graph to accelerate processing and remove or reduce the overhead of padding. This is for example supported in the PyG library. However, the sizes of small graphs can vary substantially with respect to the number of nodes and edges, and hence the size of the combined graph can still vary considerably, especially for small batch sizes. Therefore, the costs of excessive padding and wasted compute are still incurred when working with static shapes, which are preferred for maximum acceleration. This paper proposes a new hardware agnostic approach -- tuple packing -- for generating batches that cause minimal overhead. The algorithm extends recently introduced sequence packing approaches to work on the 2D tuples of (|nodes|, |edges|). A monotone heuristic is applied to the 2D histogram of tuple values to define a priority for packing histogram bins together with the objective to reach a limit on the number of nodes as well as the number of edges. Experiments verify the effectiveness of the algorithm on multiple datasets.
Classifier Transfer with Data Selection Strategies for Online Support Vector Machine Classification with Class Imbalance
Aug 10, 2022Abstract:Objective: Classifier transfers usually come with dataset shifts. To overcome them, online strategies have to be applied. For practical applications, limitations in the computational resources for the adaptation of batch learning algorithms, like the SVM, have to be considered. Approach: We review and compare several strategies for online learning with SVMs. We focus on data selection strategies which limit the size of the stored training data [...] Main Results: For different data shifts, different criteria are appropriate. For the synthetic data, adding all samples to the pool of considered samples performs often significantly worse than other criteria. Especially, adding only misclassified samples performed astoundingly well. Here, balancing criteria were very important when the other criteria were not well chosen. For the transfer setups, the results show that the best strategy depends on the intensity of the drift during the transfer. Adding all and removing the oldest samples results in the best performance, whereas for smaller drifts, it can be sufficient to only add potential new support vectors of the SVM which reduces processing resources. Significance: For BCIs based on EEG models, trained on data from a calibration session, a previous recording session, or even from a recording session with one or several other subjects, are used. This transfer of the learned model usually decreases the performance and can therefore benefit from online learning which adapts the classifier like the established SVM. We show that by using the right combination of data selection criteria, it is possible to adapt the classifier and largely increase the performance. Furthermore, in some cases it is possible to speed up the processing and save computational by updating with a subset of special samples and keeping a small subset of samples for training the classifier.
NanoBatch DPSGD: Exploring Differentially Private learning on ImageNet with low batch sizes on the IPU
Sep 24, 2021
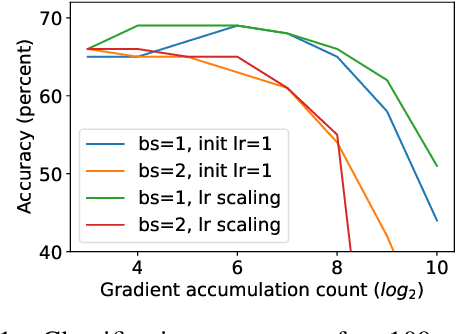
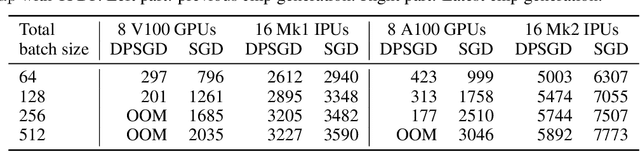
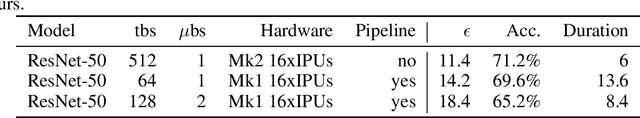
Abstract:Differentially private SGD (DPSGD) has recently shown promise in deep learning. However, compared to non-private SGD, the DPSGD algorithm places computational overheads that can undo the benefit of batching in GPUs. Microbatching is a standard method to alleviate this and is fully supported in the TensorFlow Privacy library (TFDP). However, this technique, while improving training times also reduces the quality of the gradients and degrades the classification accuracy. Recent works that for example use the JAX framework show promise in also alleviating this but still show degradation in throughput from non-private to private SGD on CNNs, and have not yet shown ImageNet implementations. In our work, we argue that low batch sizes using group normalization on ResNet-50 can yield high accuracy and privacy on Graphcore IPUs. This enables DPSGD training of ResNet-50 on ImageNet in just 6 hours (100 epochs) on an IPU-POD16 system.
Packing: Towards 2x NLP BERT Acceleration
Jun 29, 2021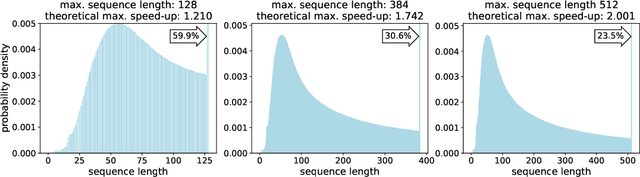
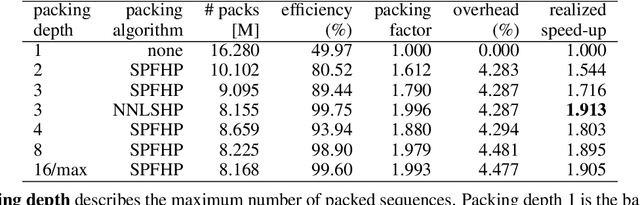


Abstract:We find that at sequence length 512 padding tokens represent in excess of 50% of the Wikipedia dataset used for pretraining BERT (Bidirectional Encoder Representations from Transformers). Therefore by removing all padding we achieve a 2x speed-up in terms of sequences/sec. To exploit this characteristic of the dataset, we develop and contrast two deterministic packing algorithms. Both algorithms rely on the assumption that sequences are interchangeable and therefore packing can be performed on the histogram of sequence lengths, rather than per sample. This transformation of the problem leads to algorithms which are fast and have linear complexity in dataset size. The shortest-pack-first histogram-packing (SPFHP) algorithm determines the packing order for the Wikipedia dataset of over 16M sequences in 0.02 seconds. The non-negative least-squares histogram-packing (NNLSHP) algorithm converges in 28.4 seconds but produces solutions which are more depth efficient, managing to get near optimal packing by combining a maximum of 3 sequences in one sample. Using the dataset with multiple sequences per sample requires additional masking in the attention layer and a modification of the MLM loss function. We demonstrate that both of these changes are straightforward to implement and have relatively little impact on the achievable performance gain on modern hardware. Finally, we pretrain BERT-Large using the packed dataset, demonstrating no loss of convergence and the desired 2x speed-up.
OrigamiSet1.0: Two New Datasets for Origami Classification and Difficulty Estimation
Jan 14, 2021
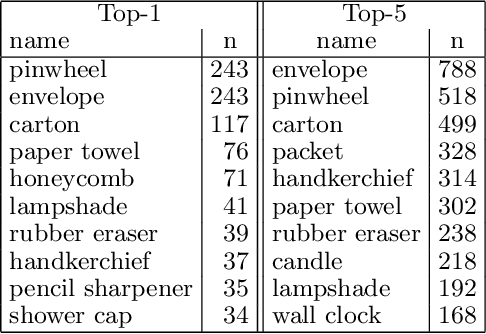
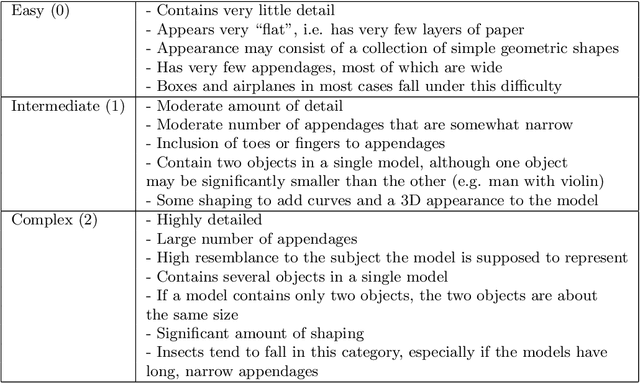
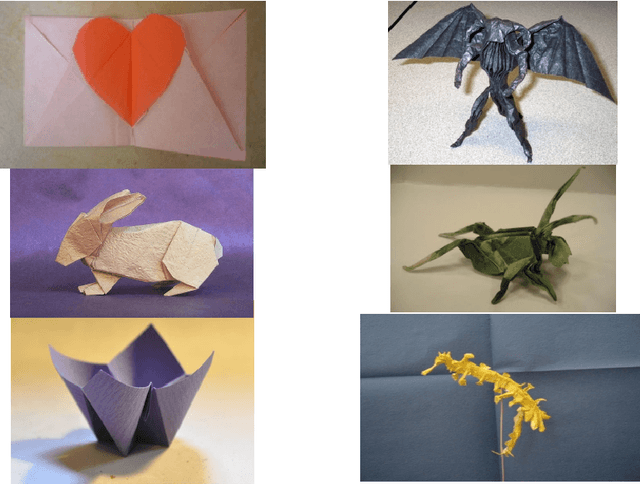
Abstract:Origami is becoming more and more relevant to research. However, there is no public dataset yet available and there hasn't been any research on this topic in machine learning. We constructed an origami dataset using images from the multimedia commons and other databases. It consists of two subsets: one for classification of origami images and the other for difficulty estimation. We obtained 16000 images for classification (half origami, half other objects) and 1509 for difficulty estimation with $3$ different categories (easy: 764, intermediate: 427, complex: 318). The data can be downloaded at: https://github.com/multimedia-berkeley/OriSet. Finally, we provide machine learning baselines.
Hardware-accelerated Simulation-based Inference of Stochastic Epidemiology Models for COVID-19
Dec 23, 2020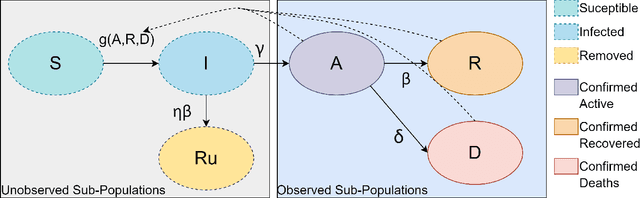
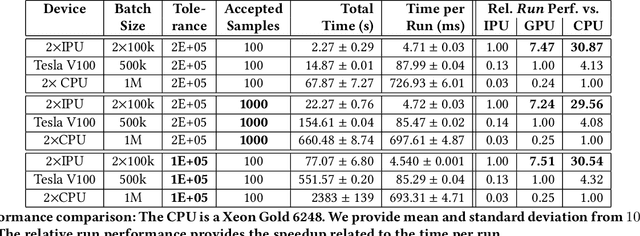
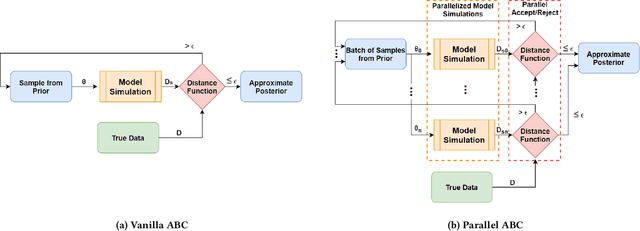
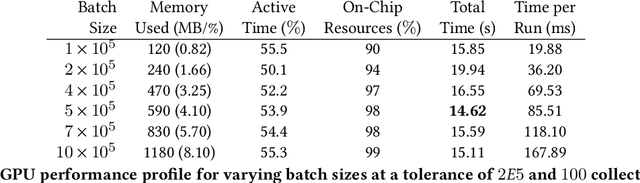
Abstract:Epidemiology models are central in understanding and controlling large scale pandemics. Several epidemiology models require simulation-based inference such as Approximate Bayesian Computation (ABC) to fit their parameters to observations. ABC inference is highly amenable to efficient hardware acceleration. In this work, we develop parallel ABC inference of a stochastic epidemiology model for COVID-19. The statistical inference framework is implemented and compared on Intel Xeon CPU, NVIDIA Tesla V100 GPU and the Graphcore Mk1 IPU, and the results are discussed in the context of their computational architectures. Results show that GPUs are 4x and IPUs are 30x faster than Xeon CPUs. Extensive performance analysis indicates that the difference between IPU and GPU can be attributed to higher communication bandwidth, closeness of memory to compute, and higher compute power in the IPU. The proposed framework scales across 16 IPUs, with scaling overhead not exceeding 8% for the experiments performed. We present an example of our framework in practice, performing inference on the epidemiology model across three countries, and giving a brief overview of the results.
A First Step Towards Distribution Invariant Regression Metrics
Sep 10, 2020


Abstract:Regression evaluation has been performed for decades. Some metrics have been identified to be robust against shifting and scaling of the data but considering the different distributions of data is much more difficult to address (imbalance problem) even though it largely impacts the comparability between evaluations on different datasets. In classification, it has been stated repeatedly that performance metrics like the F-Measure and Accuracy are highly dependent on the class distribution and that comparisons between different datasets with different distributions are impossible. We show that the same problem exists in regression. The distribution of odometry parameters in robotic applications can for example largely vary between different recording sessions. Here, we need regression algorithms that either perform equally well for all function values, or that focus on certain boundary regions like high speed. This has to be reflected in the evaluation metric. We propose the modification of established regression metrics by weighting with the inverse distribution of function values $Y$ or the samples $X$ using an automatically tuned Gaussian kernel density estimator. We show on synthetic and robotic data in reproducible experiments that classical metrics behave wrongly, whereas our new metrics are less sensitive to changing distributions, especially when correcting by the marginal distribution in $X$. Our new evaluation concept enables the comparison of results between different datasets with different distributions. Furthermore, it can reveal overfitting of a regression algorithm to overrepresented target values. As an outcome, non-overfitting regression algorithms will be more likely chosen due to our corrected metrics.
Learning of Multi-Context Models for Autonomous Underwater Vehicles
Sep 17, 2018



Abstract:Multi-context model learning is crucial for marine robotics where several factors can cause disturbances to the system's dynamics. This work addresses the problem of identifying multiple contexts of an AUV model. We build a simulation model of the robot from experimental data, and use it to fill in the missing data and generate different model contexts. We implement an architecture based on long-short-term-memory (LSTM) networks to learn the different contexts directly from the data. We show that the LSTM network can achieve high classification accuracy compared to baseline methods, showing robustness against noise and scaling efficiently on large datasets.
Generalizing, Decoding, and Optimizing Support Vector Machine Classification
Jan 15, 2018



Abstract:The classification of complex data usually requires the composition of processing steps. Here, a major challenge is the selection of optimal algorithms for preprocessing and classification (including parameterizations). Nowadays, parts of the optimization process are automized but expert knowledge and manual work are still required. We present three steps to face this process and ease the optimization. Namely, we take a theoretical view on classical classifiers, provide an approach to interpret the classifier together with the preprocessing, and integrate both into one framework which enables a semiautomatic optimization of the processing chain and which interfaces numerous algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge