Kristen W. Yeom
Federated Learning on Heterogenous Data using Chest CT
Mar 23, 2023Abstract:Large data have accelerated advances in AI. While it is well known that population differences from genetics, sex, race, diet, and various environmental factors contribute significantly to disease, AI studies in medicine have largely focused on locoregional patient cohorts with less diverse data sources. Such limitation stems from barriers to large-scale data share in medicine and ethical concerns over data privacy. Federated learning (FL) is one potential pathway for AI development that enables learning across hospitals without data share. In this study, we show the results of various FL strategies on one of the largest and most diverse COVID-19 chest CT datasets: 21 participating hospitals across five continents that comprise >10,000 patients with >1 million images. We present three techniques: Fed Averaging (FedAvg), Incremental Institutional Learning (IIL), and Cyclical Incremental Institutional Learning (CIIL). We also propose an FL strategy that leverages synthetically generated data to overcome class imbalances and data size disparities across centers. We show that FL can achieve comparable performance to Centralized Data Sharing (CDS) while maintaining high performance across sites with small, underrepresented data. We investigate the strengths and weaknesses for all technical approaches on this heterogeneous dataset including the robustness to non-Independent and identically distributed (non-IID) diversity of data. We also describe the sources of data heterogeneity such as age, sex, and site locations in the context of FL and show how even among the correctly labeled populations, disparities can arise due to these biases.
NanoBatch DPSGD: Exploring Differentially Private learning on ImageNet with low batch sizes on the IPU
Sep 24, 2021
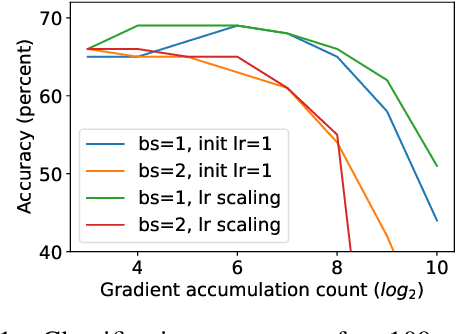
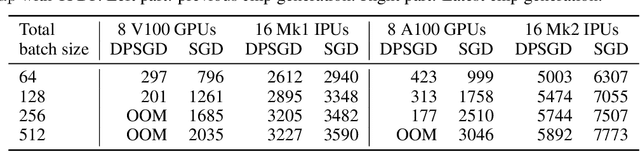
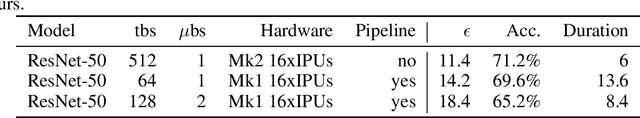
Abstract:Differentially private SGD (DPSGD) has recently shown promise in deep learning. However, compared to non-private SGD, the DPSGD algorithm places computational overheads that can undo the benefit of batching in GPUs. Microbatching is a standard method to alleviate this and is fully supported in the TensorFlow Privacy library (TFDP). However, this technique, while improving training times also reduces the quality of the gradients and degrades the classification accuracy. Recent works that for example use the JAX framework show promise in also alleviating this but still show degradation in throughput from non-private to private SGD on CNNs, and have not yet shown ImageNet implementations. In our work, we argue that low batch sizes using group normalization on ResNet-50 can yield high accuracy and privacy on Graphcore IPUs. This enables DPSGD training of ResNet-50 on ImageNet in just 6 hours (100 epochs) on an IPU-POD16 system.
Learning-based Single-step Quantitative Susceptibility Mapping Reconstruction Without Brain Extraction
May 15, 2019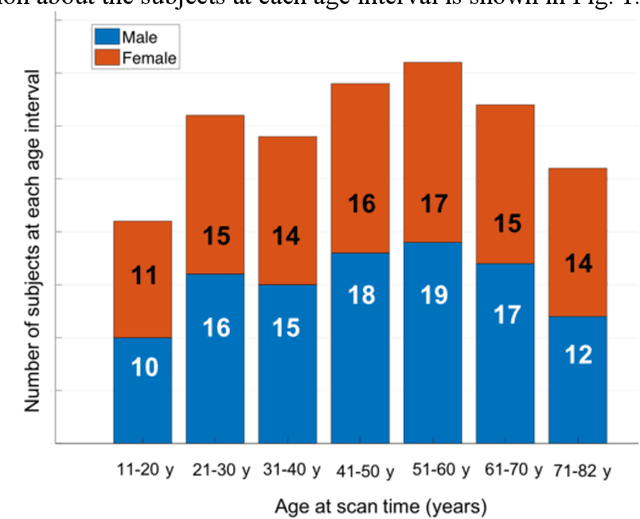
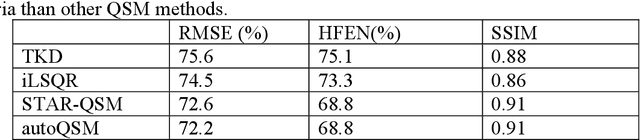
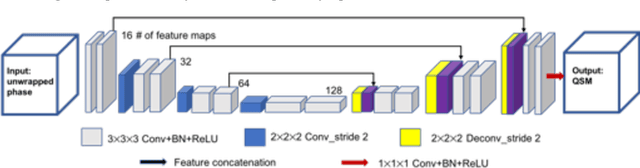
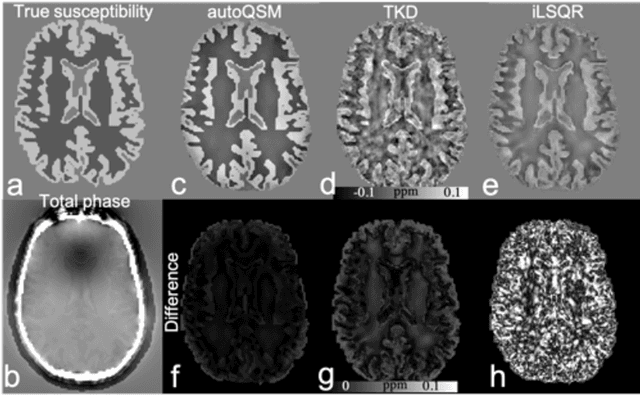
Abstract:Quantitative susceptibility mapping (QSM) estimates the underlying tissue magnetic susceptibility from MRI gradient-echo phase signal and typically requires several processing steps. These steps involve phase unwrapping, brain volume extraction, background phase removal and solving an ill-posed inverse problem. The resulting susceptibility map is known to suffer from inaccuracy near the edges of the brain tissues, in part due to imperfect brain extraction, edge erosion of the brain tissue and the lack of phase measurement outside the brain. This inaccuracy has thus hindered the application of QSM for measuring the susceptibility of tissues near the brain edges, e.g., quantifying cortical layers and generating superficial venography. To address these challenges, we propose a learning-based QSM reconstruction method that directly estimates the magnetic susceptibility from total phase images without the need for brain extraction and background phase removal, referred to as autoQSM. The neural network has a modified U-net structure and is trained using QSM maps computed by a two-step QSM method. 209 healthy subjects with ages ranging from 11 to 82 years were employed for patch-wise network training. The network was validated on data dissimilar to the training data, e.g. in vivo mouse brain data and brains with lesions, which suggests that the network has generalized and learned the underlying mathematical relationship between magnetic field perturbation and magnetic susceptibility. AutoQSM was able to recover magnetic susceptibility of anatomical structures near the edges of the brain including the veins covering the cortical surface, spinal cord and nerve tracts near the mouse brain boundaries. The advantages of high-quality maps, no need for brain volume extraction and high reconstruction speed demonstrate its potential for future applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge