Steven Cao
On the Entropy Calibration of Language Models
Nov 15, 2025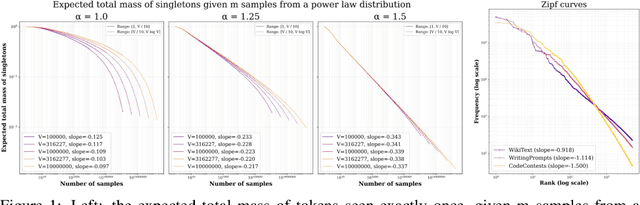
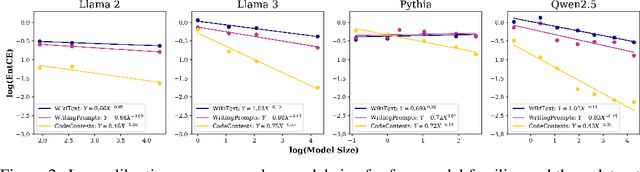
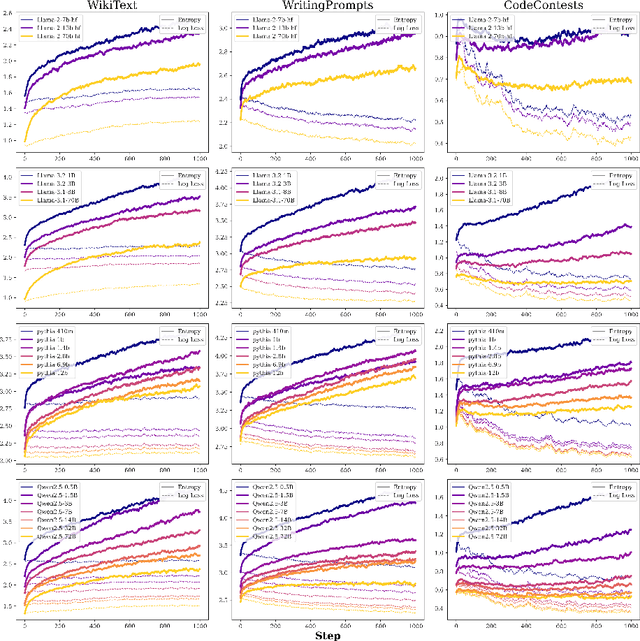
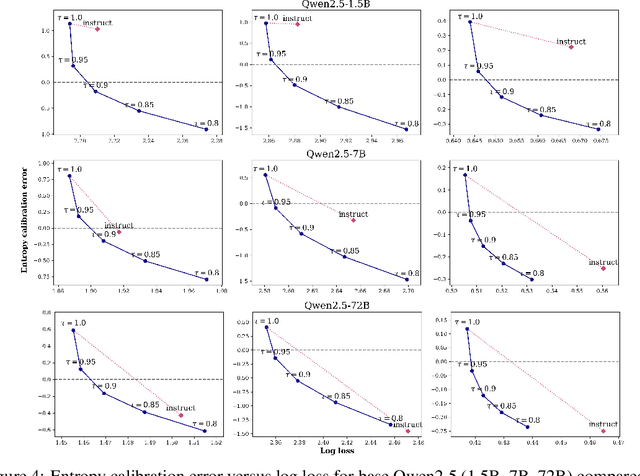
Abstract:We study the problem of entropy calibration, which asks whether a language model's entropy over generations matches its log loss on human text. Past work found that models are miscalibrated, with entropy per step increasing (and text quality decreasing) as generations grow longer. This error accumulation is a fundamental problem in autoregressive models, and the standard solution is to truncate the distribution, which improves text quality at the cost of diversity. In this paper, we ask: is miscalibration likely to improve with scale, and is it theoretically possible to calibrate without tradeoffs? To build intuition, we first study a simplified theoretical setting to characterize the scaling behavior of miscalibration with respect to dataset size. We find that the scaling behavior depends on the power law exponent of the data distribution -- in particular, for a power law exponent close to 1, the scaling exponent is close to 0, meaning that miscalibration improves very slowly with scale. Next, we measure miscalibration empirically in language models ranging from 0.5B to 70B parameters. We find that the observed scaling behavior is similar to what is predicted by the simplified setting: our fitted scaling exponents for text are close to 0, meaning that larger models accumulate error at a similar rate as smaller ones. This scaling (or, lack thereof) provides one explanation for why we sample from larger models with similar amounts of truncation as smaller models, even though the larger models are of higher quality. However, truncation is not a satisfying solution because it comes at the cost of increased log loss. In theory, is it even possible to reduce entropy while preserving log loss? We prove that it is possible, if we assume access to a black box which can fit models to predict the future entropy of text.
Reducing Intraspecies and Interspecies Covariate Shift in Traumatic Brain Injury EEG of Humans and Mice Using Transfer Euclidean Alignment
Oct 03, 2023



Abstract:While analytics of sleep electroencephalography (EEG) holds certain advantages over other methods in clinical applications, high variability across subjects poses a significant challenge when it comes to deploying machine learning models for classification tasks in the real world. In such instances, machine learning models that exhibit exceptional performance on a specific dataset may not necessarily demonstrate similar proficiency when applied to a distinct dataset for the same task. The scarcity of high-quality biomedical data further compounds this challenge, making it difficult to evaluate the model's generality comprehensively. In this paper, we introduce Transfer Euclidean Alignment - a transfer learning technique to tackle the problem of the dearth of human biomedical data for training deep learning models. We tested the robustness of this transfer learning technique on various rule-based classical machine learning models as well as the EEGNet-based deep learning model by evaluating on different datasets, including human and mouse data in a binary classification task of detecting individuals with versus without traumatic brain injury (TBI). By demonstrating notable improvements with an average increase of 14.42% for intraspecies datasets and 5.53% for interspecies datasets, our findings underscore the importance of the use of transfer learning to improve the performance of machine learning and deep learning models when using diverse datasets for training.
One-sided Matrix Completion from Two Observations Per Row
Jun 06, 2023



Abstract:Given only a few observed entries from a low-rank matrix $X$, matrix completion is the problem of imputing the missing entries, and it formalizes a wide range of real-world settings that involve estimating missing data. However, when there are too few observed entries to complete the matrix, what other aspects of the underlying matrix can be reliably recovered? We study one such problem setting, that of "one-sided" matrix completion, where our goal is to recover the right singular vectors of $X$, even in the regime where recovering the left singular vectors is impossible, which arises when there are more rows than columns and very few observations. We propose a natural algorithm that involves imputing the missing values of the matrix $X^TX$ and show that even with only two observations per row in $X$, we can provably recover $X^TX$ as long as we have at least $\Omega(r^2 d \log d)$ rows, where $r$ is the rank and $d$ is the number of columns. We evaluate our algorithm on one-sided recovery of synthetic data and low-coverage genome sequencing. In these settings, our algorithm substantially outperforms standard matrix completion and a variety of direct factorization methods.
Low-Complexity Probing via Finding Subnetworks
Apr 08, 2021



Abstract:The dominant approach in probing neural networks for linguistic properties is to train a new shallow multi-layer perceptron (MLP) on top of the model's internal representations. This approach can detect properties encoded in the model, but at the cost of adding new parameters that may learn the task directly. We instead propose a subtractive pruning-based probe, where we find an existing subnetwork that performs the linguistic task of interest. Compared to an MLP, the subnetwork probe achieves both higher accuracy on pre-trained models and lower accuracy on random models, so it is both better at finding properties of interest and worse at learning on its own. Next, by varying the complexity of each probe, we show that subnetwork probing Pareto-dominates MLP probing in that it achieves higher accuracy given any budget of probe complexity. Finally, we analyze the resulting subnetworks across various tasks to locate where each task is encoded, and we find that lower-level tasks are captured in lower layers, reproducing similar findings in past work.
Unsupervised Parsing via Constituency Tests
Oct 07, 2020
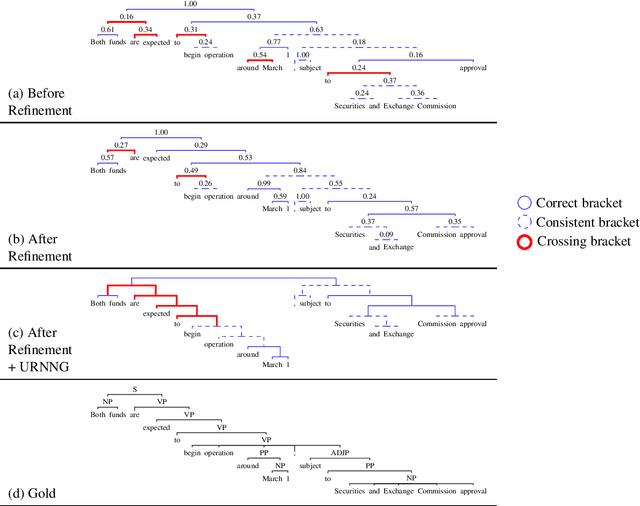

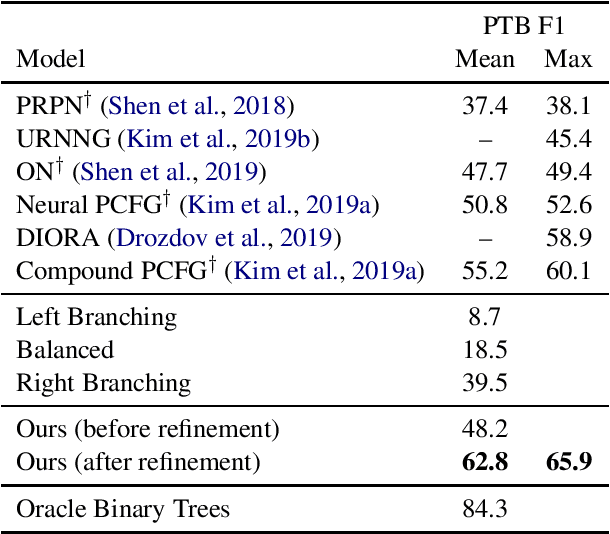
Abstract:We propose a method for unsupervised parsing based on the linguistic notion of a constituency test. One type of constituency test involves modifying the sentence via some transformation (e.g. replacing the span with a pronoun) and then judging the result (e.g. checking if it is grammatical). Motivated by this idea, we design an unsupervised parser by specifying a set of transformations and using an unsupervised neural acceptability model to make grammaticality decisions. To produce a tree given a sentence, we score each span by aggregating its constituency test judgments, and we choose the binary tree with the highest total score. While this approach already achieves performance in the range of current methods, we further improve accuracy by fine-tuning the grammaticality model through a refinement procedure, where we alternate between improving the estimated trees and improving the grammaticality model. The refined model achieves 62.8 F1 on the Penn Treebank test set, an absolute improvement of 7.6 points over the previous best published result.
Multilingual Alignment of Contextual Word Representations
Feb 12, 2020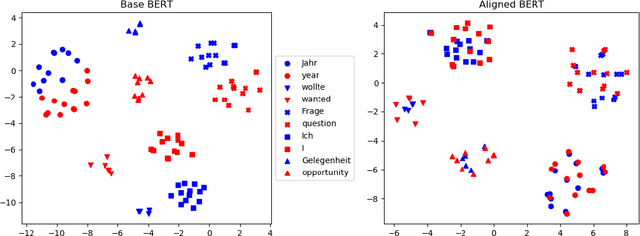
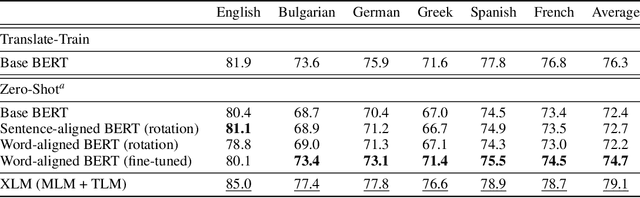
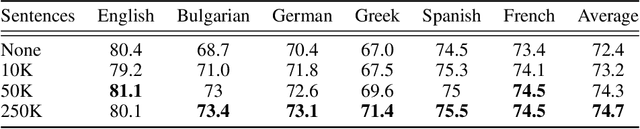
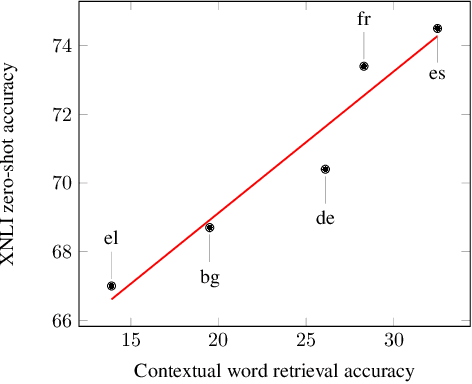
Abstract:We propose procedures for evaluating and strengthening contextual embedding alignment and show that they are useful in analyzing and improving multilingual BERT. In particular, after our proposed alignment procedure, BERT exhibits significantly improved zero-shot performance on XNLI compared to the base model, remarkably matching pseudo-fully-supervised translate-train models for Bulgarian and Greek. Further, to measure the degree of alignment, we introduce a contextual version of word retrieval and show that it correlates well with downstream zero-shot transfer. Using this word retrieval task, we also analyze BERT and find that it exhibits systematic deficiencies, e.g. worse alignment for open-class parts-of-speech and word pairs written in different scripts, that are corrected by the alignment procedure. These results support contextual alignment as a useful concept for understanding large multilingual pre-trained models.
Learning-based Single-step Quantitative Susceptibility Mapping Reconstruction Without Brain Extraction
May 15, 2019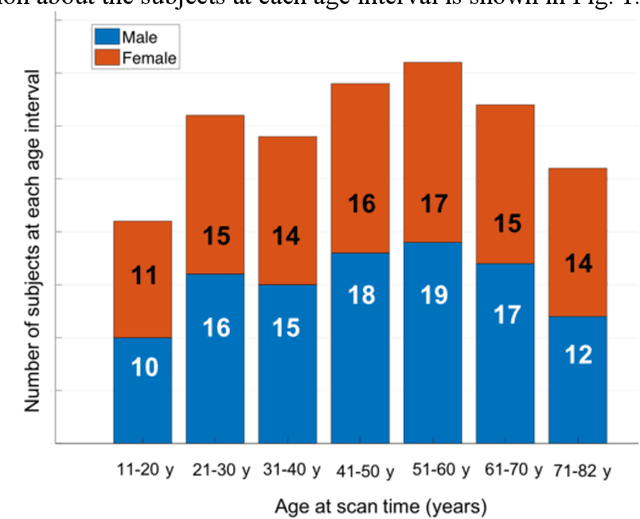
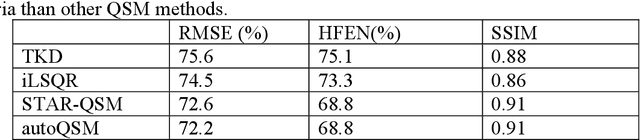
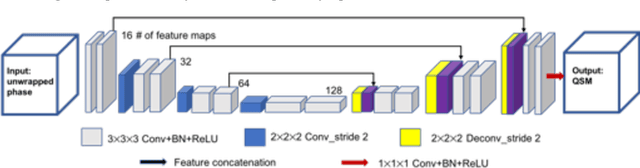
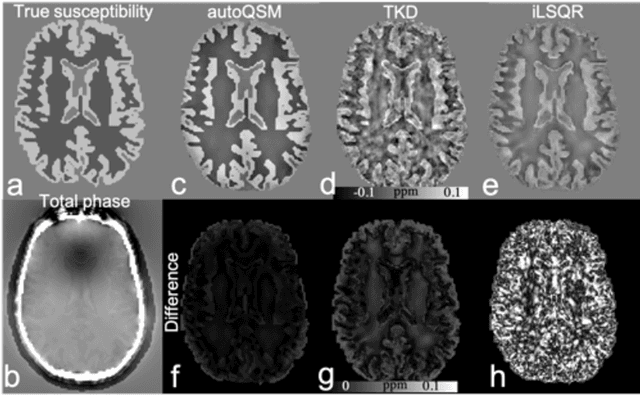
Abstract:Quantitative susceptibility mapping (QSM) estimates the underlying tissue magnetic susceptibility from MRI gradient-echo phase signal and typically requires several processing steps. These steps involve phase unwrapping, brain volume extraction, background phase removal and solving an ill-posed inverse problem. The resulting susceptibility map is known to suffer from inaccuracy near the edges of the brain tissues, in part due to imperfect brain extraction, edge erosion of the brain tissue and the lack of phase measurement outside the brain. This inaccuracy has thus hindered the application of QSM for measuring the susceptibility of tissues near the brain edges, e.g., quantifying cortical layers and generating superficial venography. To address these challenges, we propose a learning-based QSM reconstruction method that directly estimates the magnetic susceptibility from total phase images without the need for brain extraction and background phase removal, referred to as autoQSM. The neural network has a modified U-net structure and is trained using QSM maps computed by a two-step QSM method. 209 healthy subjects with ages ranging from 11 to 82 years were employed for patch-wise network training. The network was validated on data dissimilar to the training data, e.g. in vivo mouse brain data and brains with lesions, which suggests that the network has generalized and learned the underlying mathematical relationship between magnetic field perturbation and magnetic susceptibility. AutoQSM was able to recover magnetic susceptibility of anatomical structures near the edges of the brain including the veins covering the cortical surface, spinal cord and nerve tracts near the mouse brain boundaries. The advantages of high-quality maps, no need for brain volume extraction and high reconstruction speed demonstrate its potential for future applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge