Ling Zhong
Self-Correction Distillation for Structured Data Question Answering
Nov 17, 2025Abstract:Structured data question answering (QA), including table QA, Knowledge Graph (KG) QA, and temporal KG QA, is a pivotal research area. Advances in large language models (LLMs) have driven significant progress in unified structural QA frameworks like TrustUQA. However, these frameworks face challenges when applied to small-scale LLMs since small-scale LLMs are prone to errors in generating structured queries. To improve the structured data QA ability of small-scale LLMs, we propose a self-correction distillation (SCD) method. In SCD, an error prompt mechanism (EPM) is designed to detect errors and provide customized error messages during inference, and a two-stage distillation strategy is designed to transfer large-scale LLMs' query-generation and error-correction capabilities to small-scale LLM. Experiments across 5 benchmarks with 3 structured data types demonstrate that our SCD achieves the best performance and superior generalization on small-scale LLM (8B) compared to other distillation methods, and closely approaches the performance of GPT4 on some datasets. Furthermore, large-scale LLMs equipped with EPM surpass the state-of-the-art results on most datasets.
Thinker: Training LLMs in Hierarchical Thinking for Deep Search via Multi-Turn Interaction
Nov 14, 2025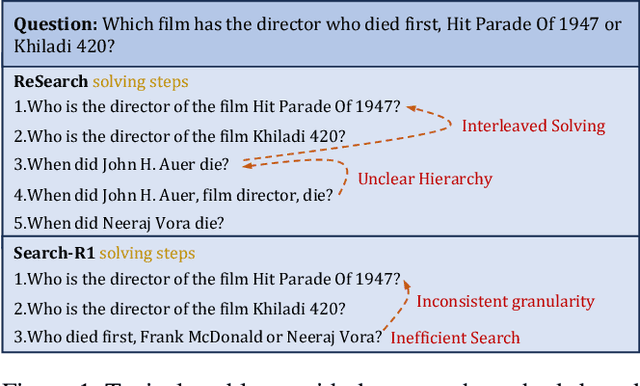
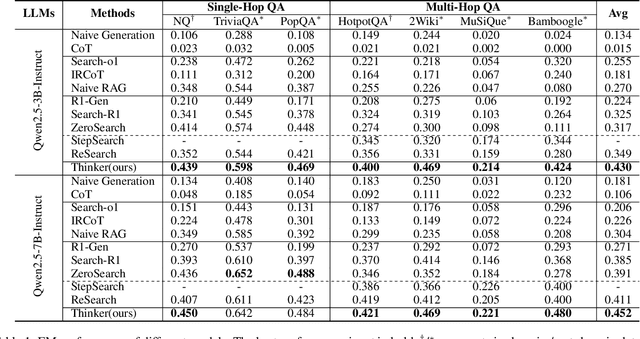
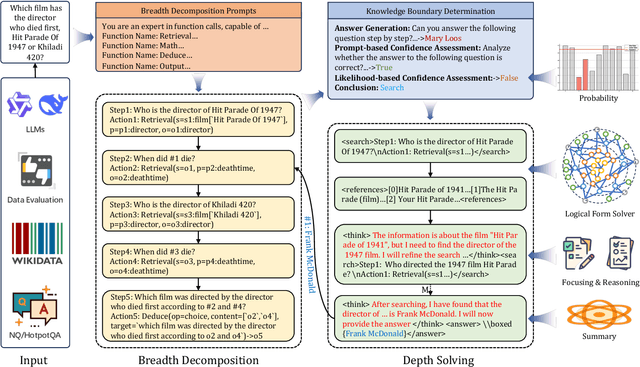
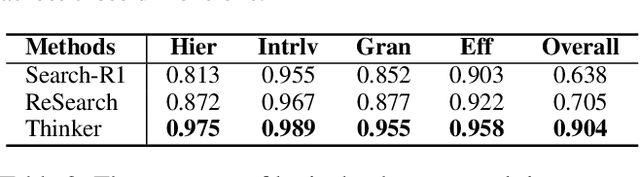
Abstract:Efficient retrieval of external knowledge bases and web pages is crucial for enhancing the reasoning abilities of LLMs. Previous works on training LLMs to leverage external retrievers for solving complex problems have predominantly employed end-to-end reinforcement learning. However, these approaches neglect supervision over the reasoning process, making it difficult to guarantee logical coherence and rigor. To address these limitations, we propose Thinker, a hierarchical thinking model for deep search through multi-turn interaction, making the reasoning process supervisable and verifiable. It decomposes complex problems into independently solvable sub-problems, each dually represented in both natural language and an equivalent logical function to support knowledge base and web searches. Concurrently, dependencies between sub-problems are passed as parameters via these logical functions, enhancing the logical coherence of the problem-solving process. To avoid unnecessary external searches, we perform knowledge boundary determination to check if a sub-problem is within the LLM's intrinsic knowledge, allowing it to answer directly. Experimental results indicate that with as few as several hundred training samples, the performance of Thinker is competitive with established baselines. Furthermore, when scaled to the full training set, Thinker significantly outperforms these methods across various datasets and model sizes. The source code is available at https://github.com/OpenSPG/KAG-Thinker.
DomainCQA: Crafting Expert-Level QA from Domain-Specific Charts
Mar 25, 2025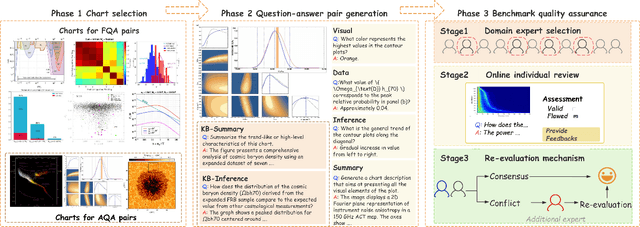
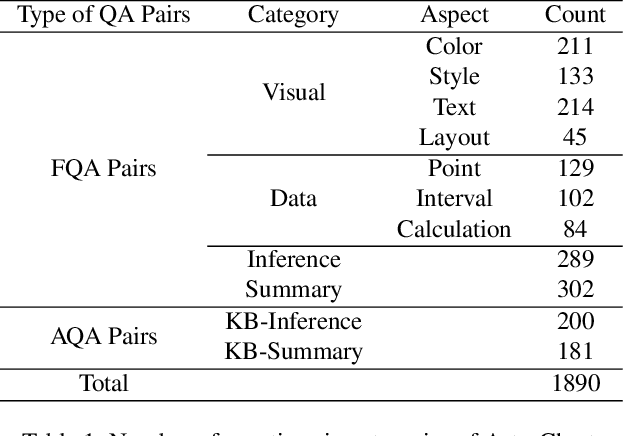
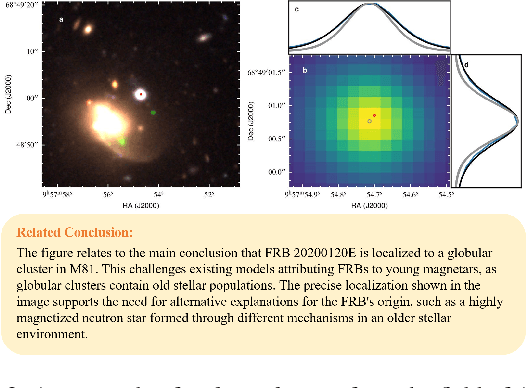
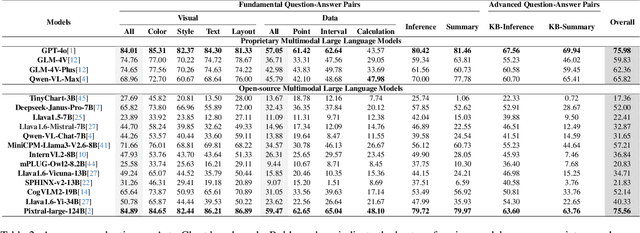
Abstract:Chart Question Answering (CQA) benchmarks are essential for evaluating the capability of Multimodal Large Language Models (MLLMs) to interpret visual data. However, current benchmarks focus primarily on the evaluation of general-purpose CQA but fail to adequately capture domain-specific challenges. We introduce DomainCQA, a systematic methodology for constructing domain-specific CQA benchmarks, and demonstrate its effectiveness by developing AstroChart, a CQA benchmark in the field of astronomy. Our evaluation shows that chart reasoning and combining chart information with domain knowledge for deeper analysis and summarization, rather than domain-specific knowledge, pose the primary challenge for existing MLLMs, highlighting a critical gap in current benchmarks. By providing a scalable and rigorous framework, DomainCQA enables more precise assessment and improvement of MLLMs for domain-specific applications.
Behavior Modeling Space Reconstruction for E-Commerce Search
Jan 30, 2025



Abstract:Delivering superior search services is crucial for enhancing customer experience and driving revenue growth. Conventionally, search systems model user behaviors by combining user preference and query item relevance statically, often through a fixed logical 'and' relationship. This paper reexamines existing approaches through a unified lens using both causal graphs and Venn diagrams, uncovering two prevalent yet significant issues: entangled preference and relevance effects, and a collapsed modeling space. To surmount these challenges, our research introduces a novel framework, DRP, which enhances search accuracy through two components to reconstruct the behavior modeling space. Specifically, we implement preference editing to proactively remove the relevance effect from preference predictions, yielding untainted user preferences. Additionally, we employ adaptive fusion, which dynamically adjusts fusion criteria to align with the varying patterns of relevance and preference, facilitating more nuanced and tailored behavior predictions within the reconstructed modeling space. Empirical validation on two public datasets and a proprietary search dataset underscores the superiority of our proposed methodology, demonstrating marked improvements in performance over existing approaches.
Efficient Knowledge Infusion via KG-LLM Alignment
Jun 06, 2024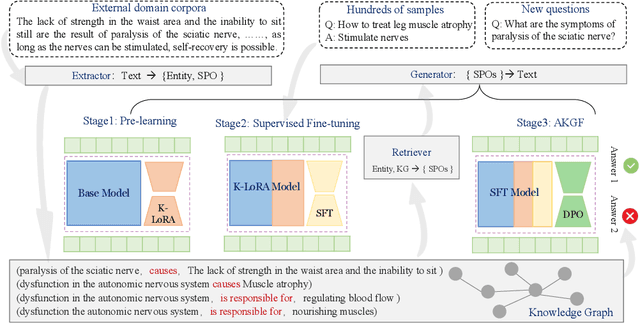
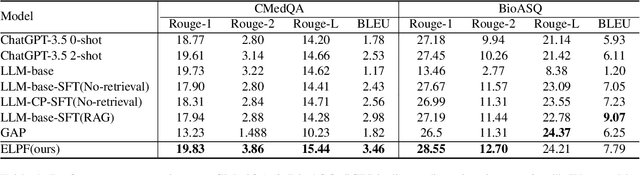
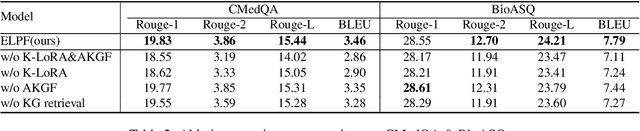
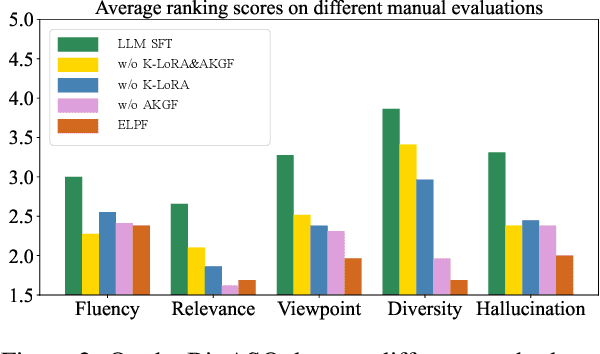
Abstract:To tackle the problem of domain-specific knowledge scarcity within large language models (LLMs), knowledge graph-retrievalaugmented method has been proven to be an effective and efficient technique for knowledge infusion. However, existing approaches face two primary challenges: knowledge mismatch between public available knowledge graphs and the specific domain of the task at hand, and poor information compliance of LLMs with knowledge graphs. In this paper, we leverage a small set of labeled samples and a large-scale corpus to efficiently construct domain-specific knowledge graphs by an LLM, addressing the issue of knowledge mismatch. Additionally, we propose a three-stage KG-LLM alignment strategyto enhance the LLM's capability to utilize information from knowledge graphs. We conduct experiments with a limited-sample setting on two biomedical question-answering datasets, and the results demonstrate that our approach outperforms existing baselines.
Effective Classification of MicroRNA Precursors Using Combinatorial Feature Mining and AdaBoost Algorithms
Oct 06, 2016



Abstract:MicroRNAs (miRNAs) are non-coding RNAs with approximately 22 nucleotides (nt) that are derived from precursor molecules. These precursor molecules or pre-miRNAs often fold into stem-loop hairpin structures. However, a large number of sequences with pre-miRNA-like hairpins can be found in genomes. It is a challenge to distinguish the real pre-miRNAs from other hairpin sequences with similar stem-loops (referred to as pseudo pre-miRNAs). Several computational methods have been developed to tackle this challenge. In this paper we propose a new method, called MirID, for identifying and classifying microRNA precursors. We collect 74 features from the sequences and secondary structures of pre-miRNAs; some of these features are taken from our previous studies on non-coding RNA prediction while others were suggested in the literature. We develop a combinatorial feature mining algorithm to identify suitable feature sets. These feature sets are then used to train support vector machines to obtain classification models, based on which classifier ensemble is constructed. Finally we use an AdaBoost algorithm to further enhance the accuracy of the classifier ensemble. Experimental results on a variety of species demonstrate the good performance of the proposed method, and its superiority over existing tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge