Julia Dietlmeier
VLSM-Ensemble: Ensembling CLIP-based Vision-Language Models for Enhanced Medical Image Segmentation
Sep 05, 2025Abstract:Vision-language models and their adaptations to image segmentation tasks present enormous potential for producing highly accurate and interpretable results. However, implementations based on CLIP and BiomedCLIP are still lagging behind more sophisticated architectures such as CRIS. In this work, instead of focusing on text prompt engineering as is the norm, we attempt to narrow this gap by showing how to ensemble vision-language segmentation models (VLSMs) with a low-complexity CNN. By doing so, we achieve a significant Dice score improvement of 6.3% on the BKAI polyp dataset using the ensembled BiomedCLIPSeg, while other datasets exhibit gains ranging from 1% to 6%. Furthermore, we provide initial results on additional four radiology and non-radiology datasets. We conclude that ensembling works differently across these datasets (from outperforming to underperforming the CRIS model), indicating a topic for future investigation by the community. The code is available at https://github.com/juliadietlmeier/VLSM-Ensemble.
Parameter-Free Bio-Inspired Channel Attention for Enhanced Cardiac MRI Reconstruction
May 29, 2025Abstract:Attention is a fundamental component of the human visual recognition system. The inclusion of attention in a convolutional neural network amplifies relevant visual features and suppresses the less important ones. Integrating attention mechanisms into convolutional neural networks enhances model performance and interpretability. Spatial and channel attention mechanisms have shown significant advantages across many downstream tasks in medical imaging. While existing attention modules have proven to be effective, their design often lacks a robust theoretical underpinning. In this study, we address this gap by proposing a non-linear attention architecture for cardiac MRI reconstruction and hypothesize that insights from ecological principles can guide the development of effective and efficient attention mechanisms. Specifically, we investigate a non-linear ecological difference equation that describes single-species population growth to devise a parameter-free attention module surpassing current state-of-the-art parameter-free methods.
Accelerating Cardiac MRI Reconstruction with CMRatt: An Attention-Driven Approach
Apr 10, 2024

Abstract:Cine cardiac magnetic resonance (CMR) imaging is recognised as the benchmark modality for the comprehensive assessment of cardiac function. Nevertheless, the acquisition process of cine CMR is considered as an impediment due to its prolonged scanning time. One commonly used strategy to expedite the acquisition process is through k-space undersampling, though it comes with a drawback of introducing aliasing effects in the reconstructed image. Lately, deep learning-based methods have shown remarkable results over traditional approaches in rapidly achieving precise CMR reconstructed images. This study aims to explore the untapped potential of attention mechanisms incorporated with a deep learning model within the context of the CMR reconstruction problem. We are motivated by the fact that attention has proven beneficial in downstream tasks such as image classification and segmentation, but has not been systematically analysed in the context of CMR reconstruction. Our primary goal is to identify the strengths and potential limitations of attention algorithms when integrated with a convolutional backbone model such as a U-Net. To achieve this, we benchmark different state-of-the-art spatial and channel attention mechanisms on the CMRxRecon dataset and quantitatively evaluate the quality of reconstruction using objective metrics. Furthermore, inspired by the best performing attention mechanism, we propose a new, simple yet effective, attention pipeline specifically optimised for the task of cardiac image reconstruction that outperforms other state-of-the-art attention methods. The layer and model code will be made publicly available.
Test-Time Adaptation with SaLIP: A Cascade of SAM and CLIP for Zero shot Medical Image Segmentation
Apr 09, 2024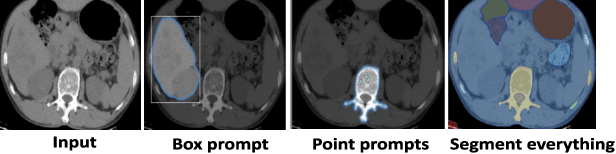
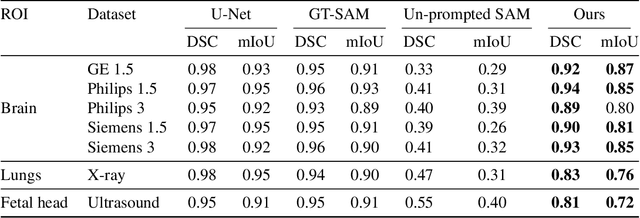
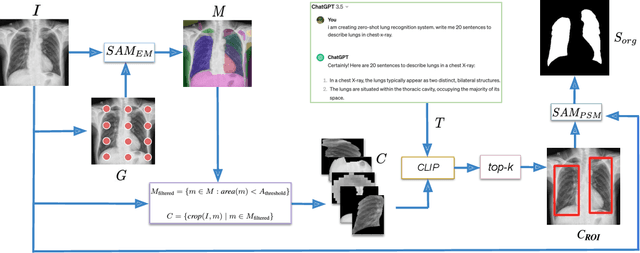

Abstract:The Segment Anything Model (SAM) and CLIP are remarkable vision foundation models (VFMs). SAM, a prompt driven segmentation model, excels in segmentation tasks across diverse domains, while CLIP is renowned for its zero shot recognition capabilities. However, their unified potential has not yet been explored in medical image segmentation. To adapt SAM to medical imaging, existing methods primarily rely on tuning strategies that require extensive data or prior prompts tailored to the specific task, making it particularly challenging when only a limited number of data samples are available. This work presents an in depth exploration of integrating SAM and CLIP into a unified framework for medical image segmentation. Specifically, we propose a simple unified framework, SaLIP, for organ segmentation. Initially, SAM is used for part based segmentation within the image, followed by CLIP to retrieve the mask corresponding to the region of interest (ROI) from the pool of SAM generated masks. Finally, SAM is prompted by the retrieved ROI to segment a specific organ. Thus, SaLIP is training and fine tuning free and does not rely on domain expertise or labeled data for prompt engineering. Our method shows substantial enhancements in zero shot segmentation, showcasing notable improvements in DICE scores across diverse segmentation tasks like brain (63.46%), lung (50.11%), and fetal head (30.82%), when compared to un prompted SAM. Code and text prompts will be available online.
The state-of-the-art in Cardiac MRI Reconstruction: Results of the CMRxRecon Challenge in MICCAI 2023
Apr 01, 2024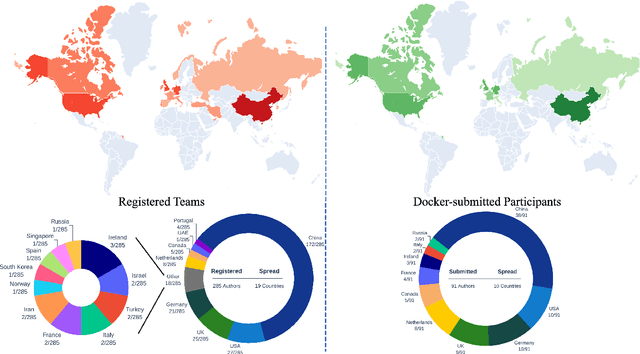
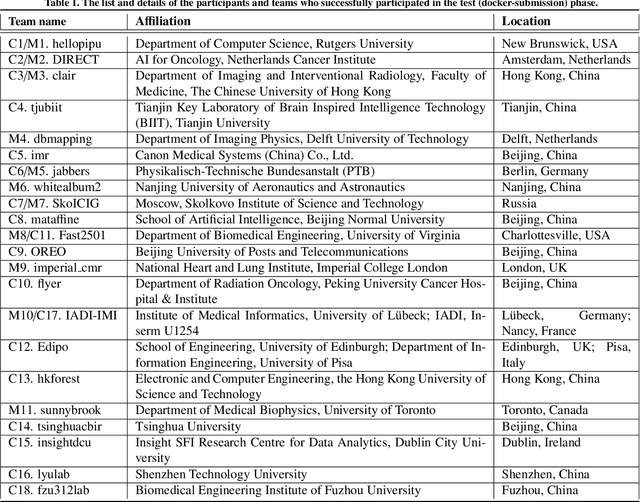
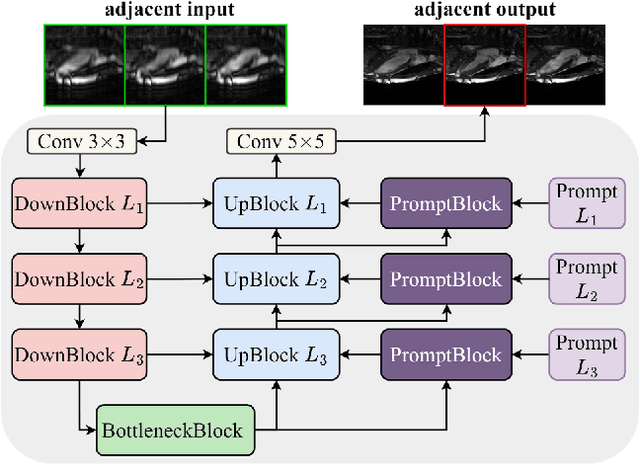
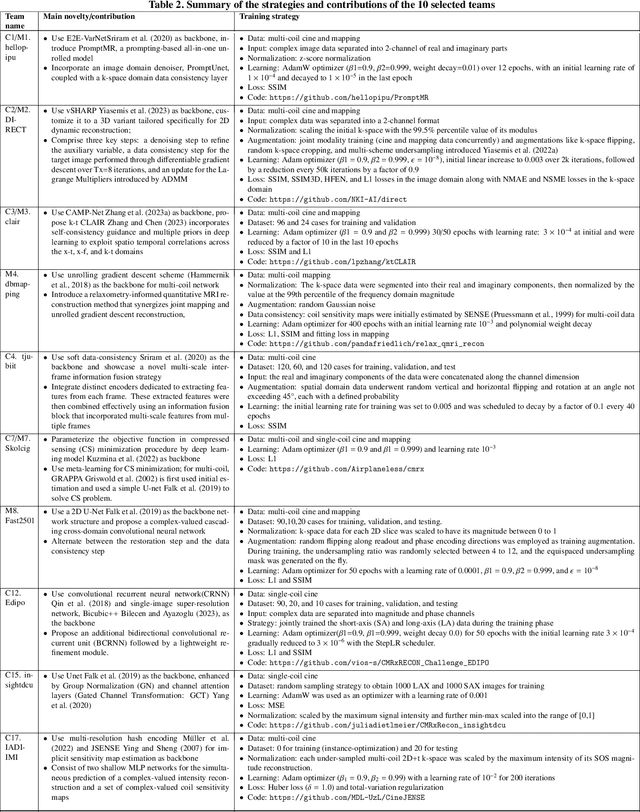
Abstract:Cardiac MRI, crucial for evaluating heart structure and function, faces limitations like slow imaging and motion artifacts. Undersampling reconstruction, especially data-driven algorithms, has emerged as a promising solution to accelerate scans and enhance imaging performance using highly under-sampled data. Nevertheless, the scarcity of publicly available cardiac k-space datasets and evaluation platform hinder the development of data-driven reconstruction algorithms. To address this issue, we organized the Cardiac MRI Reconstruction Challenge (CMRxRecon) in 2023, in collaboration with the 26th International Conference on MICCAI. CMRxRecon presented an extensive k-space dataset comprising cine and mapping raw data, accompanied by detailed annotations of cardiac anatomical structures. With overwhelming participation, the challenge attracted more than 285 teams and over 600 participants. Among them, 22 teams successfully submitted Docker containers for the testing phase, with 7 teams submitted for both cine and mapping tasks. All teams use deep learning based approaches, indicating that deep learning has predominately become a promising solution for the problem. The first-place winner of both tasks utilizes the E2E-VarNet architecture as backbones. In contrast, U-Net is still the most popular backbone for both multi-coil and single-coil reconstructions. This paper provides a comprehensive overview of the challenge design, presents a summary of the submitted results, reviews the employed methods, and offers an in-depth discussion that aims to inspire future advancements in cardiac MRI reconstruction models. The summary emphasizes the effective strategies observed in Cardiac MRI reconstruction, including backbone architecture, loss function, pre-processing techniques, physical modeling, and model complexity, thereby providing valuable insights for further developments in this field.
ConvLoRA and AdaBN based Domain Adaptation via Self-Training
Feb 07, 2024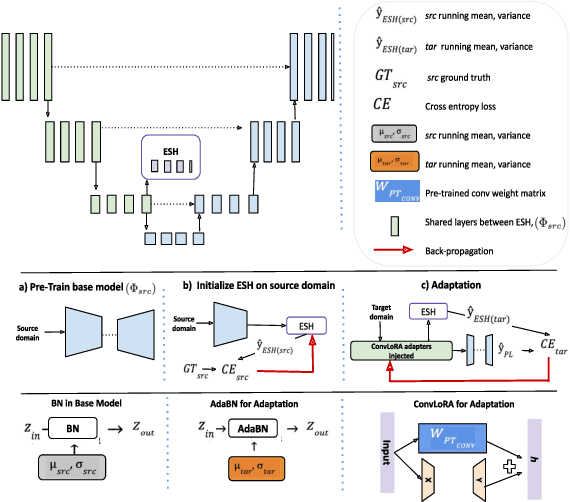
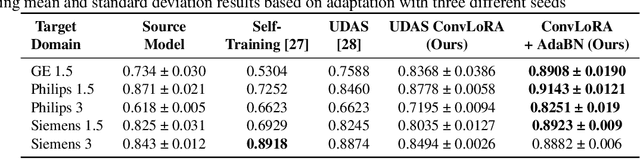
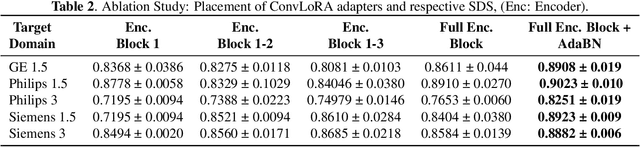
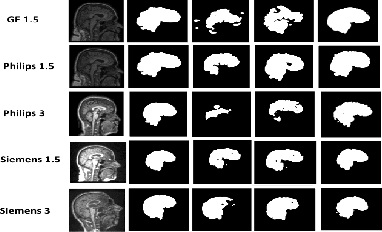
Abstract:Existing domain adaptation (DA) methods often involve pre-training on the source domain and fine-tuning on the target domain. For multi-target domain adaptation, having a dedicated/separate fine-tuned network for each target domain, that retain all the pre-trained model parameters, is prohibitively expensive. To address this limitation, we propose Convolutional Low-Rank Adaptation (ConvLoRA). ConvLoRA freezes pre-trained model weights, adds trainable low-rank decomposition matrices to convolutional layers, and backpropagates the gradient through these matrices thus greatly reducing the number of trainable parameters. To further boost adaptation, we utilize Adaptive Batch Normalization (AdaBN) which computes target-specific running statistics and use it along with ConvLoRA. Our method has fewer trainable parameters and performs better or on-par with large independent fine-tuned networks (with less than 0.9% trainable parameters of the total base model) when tested on the segmentation of Calgary-Campinas dataset containing brain MRI images. Our approach is simple, yet effective and can be applied to any deep learning-based architecture which uses convolutional and batch normalization layers. Code is available at: https://github.com/aleemsidra/ConvLoRA.
An L2-Normalized Spatial Attention Network For Accurate And Fast Classification Of Brain Tumors In 2D T1-Weighted CE-MRI Images
Aug 01, 2023


Abstract:We propose an accurate and fast classification network for classification of brain tumors in MRI images that outperforms all lightweight methods investigated in terms of accuracy. We test our model on a challenging 2D T1-weighted CE-MRI dataset containing three types of brain tumors: Meningioma, Glioma and Pituitary. We introduce an l2-normalized spatial attention mechanism that acts as a regularizer against overfitting during training. We compare our results against the state-of-the-art on this dataset and show that by integrating l2-normalized spatial attention into a baseline network we achieve a performance gain of 1.79 percentage points. Even better accuracy can be attained by combining our model in an ensemble with the pretrained VGG16 at the expense of execution speed. Our code is publicly available at https://github.com/juliadietlmeier/MRI_image_classification
Motion Aware Self-Supervision for Generic Event Boundary Detection
Oct 12, 2022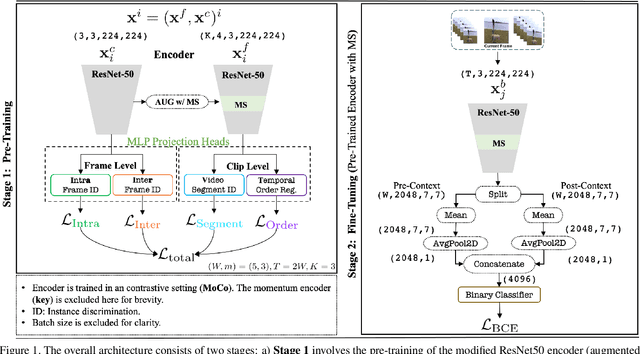
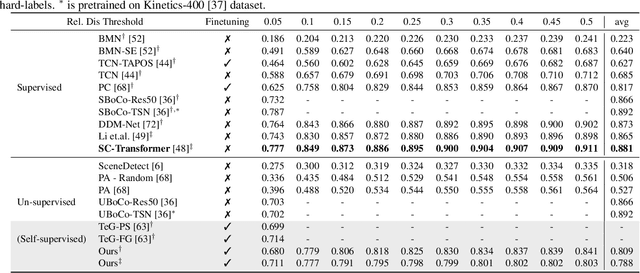
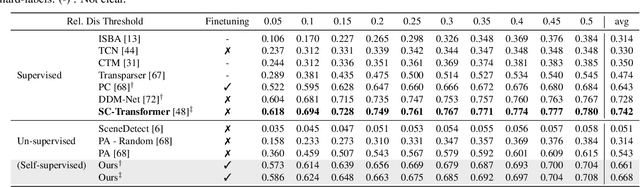
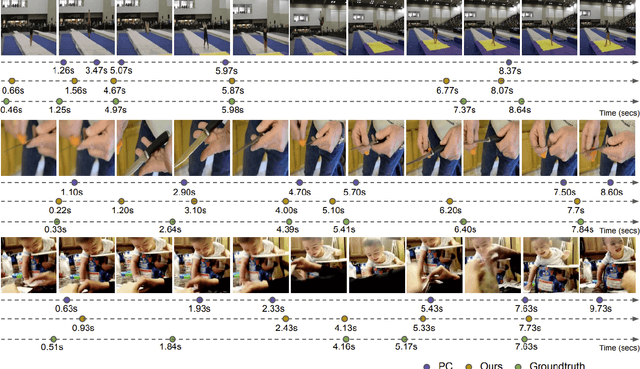
Abstract:The task of Generic Event Boundary Detection (GEBD) aims to detect moments in videos that are naturally perceived by humans as generic and taxonomy-free event boundaries. Modeling the dynamically evolving temporal and spatial changes in a video makes GEBD a difficult problem to solve. Existing approaches involve very complex and sophisticated pipelines in terms of architectural design choices, hence creating a need for more straightforward and simplified approaches. In this work, we address this issue by revisiting a simple and effective self-supervised method and augment it with a differentiable motion feature learning module to tackle the spatial and temporal diversities in the GEBD task. We perform extensive experiments on the challenging Kinetics-GEBD and TAPOS datasets to demonstrate the efficacy of the proposed approach compared to the other self-supervised state-of-the-art methods. We also show that this simple self-supervised approach learns motion features without any explicit motion-specific pretext task.
Improving Person Re-Identification with Temporal Constraints
Nov 17, 2021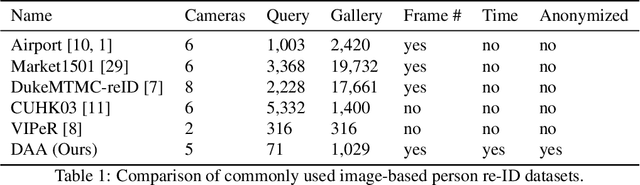

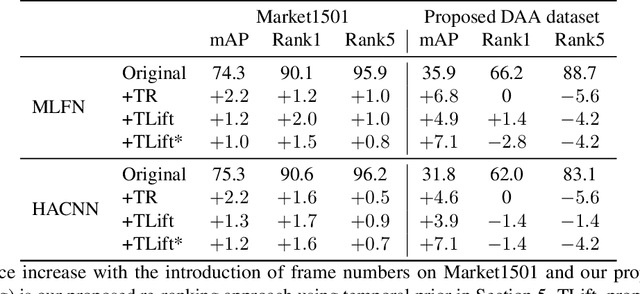
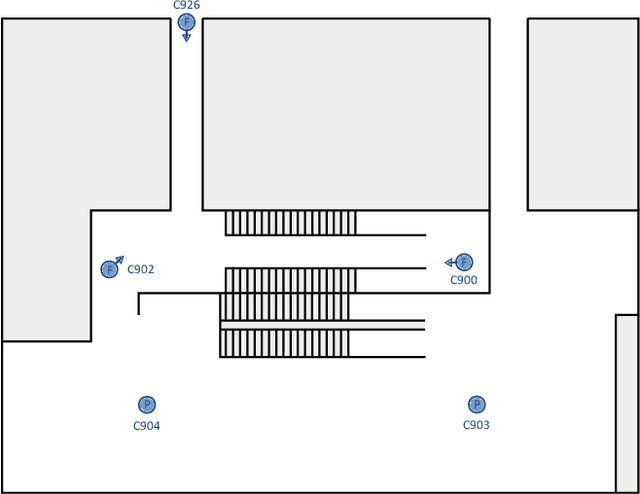
Abstract:In this paper we introduce an image-based person re-identification dataset collected across five non-overlapping camera views in the large and busy airport in Dublin, Ireland. Unlike all publicly available image-based datasets, our dataset contains timestamp information in addition to frame number, and camera and person IDs. Also our dataset has been fully anonymized to comply with modern data privacy regulations. We apply state-of-the-art person re-identification models to our dataset and show that by leveraging the available timestamp information we are able to achieve a significant gain of 37.43% in mAP and a gain of 30.22% in Rank1 accuracy. We also propose a Bayesian temporal re-ranking post-processing step, which further adds a 10.03% gain in mAP and 9.95% gain in Rank1 accuracy metrics. This work on combining visual and temporal information is not possible on other image-based person re-identification datasets. We believe that the proposed new dataset will enable further development of person re-identification research for challenging real-world applications. DAA dataset can be downloaded from https://bit.ly/3AtXTd6
Discerning Generic Event Boundaries in Long-Form Wild Videos
Jun 18, 2021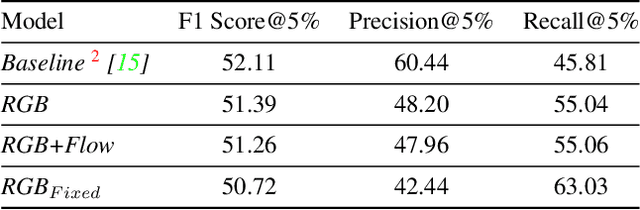
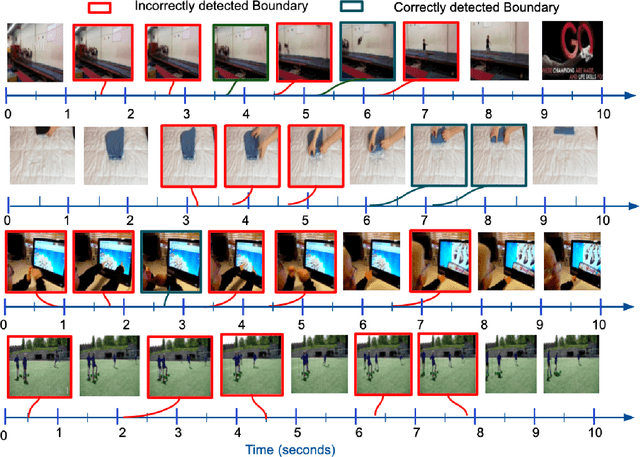
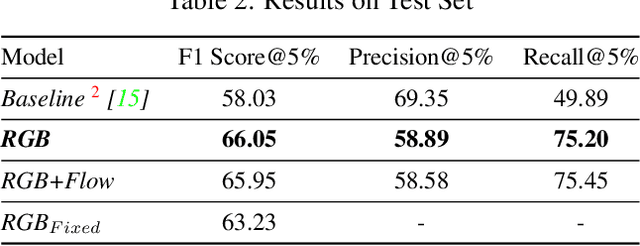
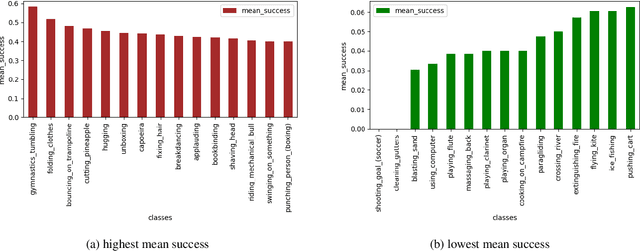
Abstract:Detecting generic, taxonomy-free event boundaries invideos represents a major stride forward towards holisticvideo understanding. In this paper we present a technique forgeneric event boundary detection based on a two stream in-flated 3D convolutions architecture, which can learn spatio-temporal features from videos. Our work is inspired from theGeneric Event Boundary Detection Challenge (part of CVPR2021 Long Form Video Understanding- LOVEU Workshop).Throughout the paper we provide an in-depth analysis ofthe experiments performed along with an interpretation ofthe results obtained.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge