Noel E. OConnor
Evaluating Image-Based Face and Eye Tracking with Event Cameras
Aug 19, 2024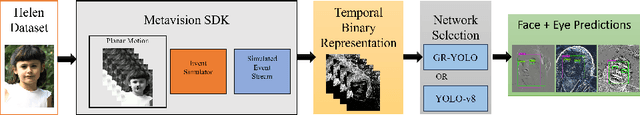
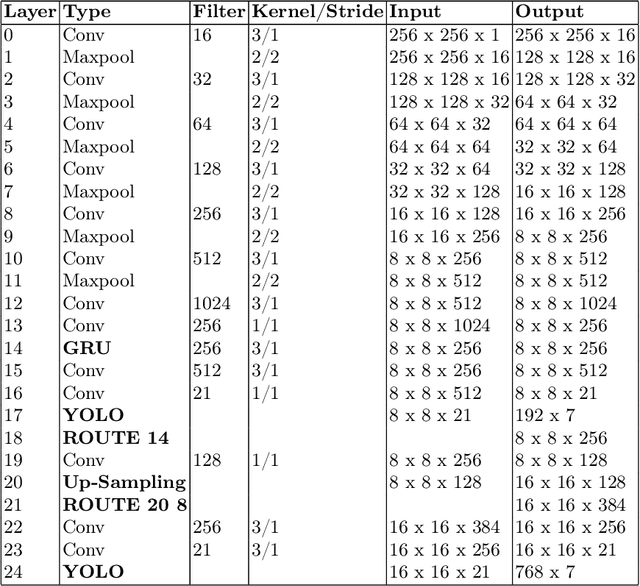


Abstract:Event Cameras, also known as Neuromorphic sensors, capture changes in local light intensity at the pixel level, producing asynchronously generated data termed ``events''. This distinct data format mitigates common issues observed in conventional cameras, like under-sampling when capturing fast-moving objects, thereby preserving critical information that might otherwise be lost. However, leveraging this data often necessitates the development of specialized, handcrafted event representations that can integrate seamlessly with conventional Convolutional Neural Networks (CNNs), considering the unique attributes of event data. In this study, We evaluate event-based Face and Eye tracking. The core objective of our study is to showcase the viability of integrating conventional algorithms with event-based data, transformed into a frame format while preserving the unique benefits of event cameras. To validate our approach, we constructed a frame-based event dataset by simulating events between RGB frames derived from the publicly accessible Helen Dataset. We assess its utility for face and eye detection tasks through the application of GR-YOLO -- a pioneering technique derived from YOLOv3. This evaluation includes a comparative analysis with results derived from training the dataset with YOLOv8. Subsequently, the trained models were tested on real event streams from various iterations of Prophesee's event cameras and further evaluated on the Faces in Event Stream (FES) benchmark dataset. The models trained on our dataset shows a good prediction performance across all the datasets obtained for validation with the best results of a mean Average precision score of 0.91. Additionally, The models trained demonstrated robust performance on real event camera data under varying light conditions.
An L2-Normalized Spatial Attention Network For Accurate And Fast Classification Of Brain Tumors In 2D T1-Weighted CE-MRI Images
Aug 01, 2023


Abstract:We propose an accurate and fast classification network for classification of brain tumors in MRI images that outperforms all lightweight methods investigated in terms of accuracy. We test our model on a challenging 2D T1-weighted CE-MRI dataset containing three types of brain tumors: Meningioma, Glioma and Pituitary. We introduce an l2-normalized spatial attention mechanism that acts as a regularizer against overfitting during training. We compare our results against the state-of-the-art on this dataset and show that by integrating l2-normalized spatial attention into a baseline network we achieve a performance gain of 1.79 percentage points. Even better accuracy can be attained by combining our model in an ensemble with the pretrained VGG16 at the expense of execution speed. Our code is publicly available at https://github.com/juliadietlmeier/MRI_image_classification
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge