Fangyijie Wang
Entropy-Guided Agreement-Diversity: A Semi-Supervised Active Learning Framework for Fetal Head Segmentation in Ultrasound
Jan 24, 2026Abstract:Fetal ultrasound (US) data is often limited due to privacy and regulatory restrictions, posing challenges for training deep learning (DL) models. While semi-supervised learning (SSL) is commonly used for fetal US image analysis, existing SSL methods typically rely on random limited selection, which can lead to suboptimal model performance by overfitting to homogeneous labeled data. To address this, we propose a two-stage Active Learning (AL) sampler, Entropy-Guided Agreement-Diversity (EGAD), for fetal head segmentation. Our method first selects the most uncertain samples using predictive entropy, and then refines the final selection using the agreement-diversity score combining cosine similarity and mutual information. Additionally, our SSL framework employs a consistency learning strategy with feature downsampling to further enhance segmentation performance. In experiments, SSL-EGAD achieves an average Dice score of 94.57\% and 96.32\% on two public datasets for fetal head segmentation, using 5\% and 10\% labeled data for training, respectively. Our method outperforms current SSL models and showcases consistent robustness across diverse pregnancy stage data. The code is available on \href{https://github.com/13204942/Semi-supervised-EGAD}{GitHub}.
PSFHS Challenge Report: Pubic Symphysis and Fetal Head Segmentation from Intrapartum Ultrasound Images
Sep 17, 2024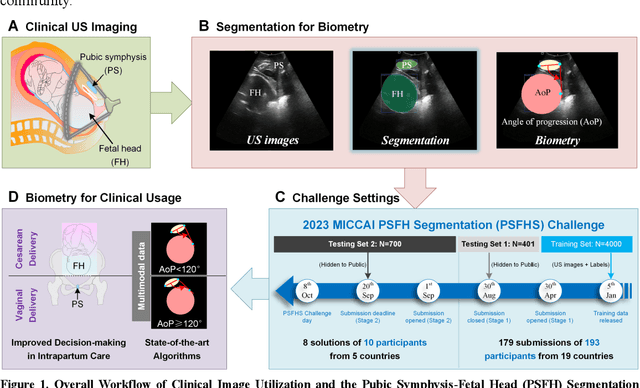
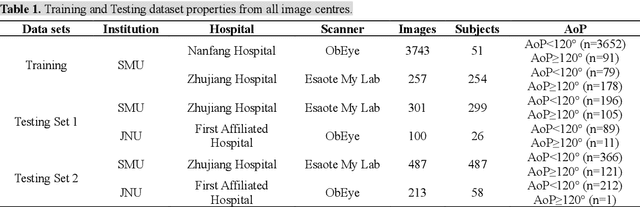
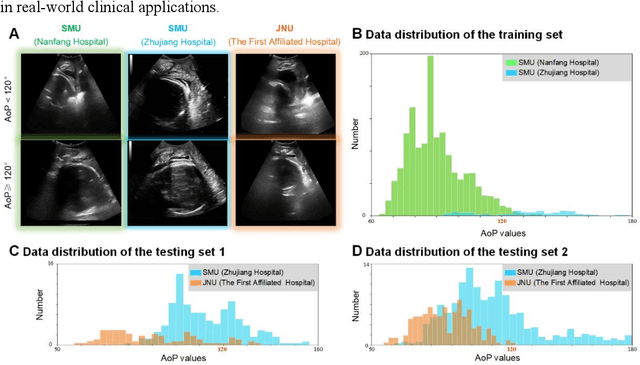

Abstract:Segmentation of the fetal and maternal structures, particularly intrapartum ultrasound imaging as advocated by the International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) for monitoring labor progression, is a crucial first step for quantitative diagnosis and clinical decision-making. This requires specialized analysis by obstetrics professionals, in a task that i) is highly time- and cost-consuming and ii) often yields inconsistent results. The utility of automatic segmentation algorithms for biometry has been proven, though existing results remain suboptimal. To push forward advancements in this area, the Grand Challenge on Pubic Symphysis-Fetal Head Segmentation (PSFHS) was held alongside the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to enhance the development of automatic segmentation algorithms at an international scale, providing the largest dataset to date with 5,101 intrapartum ultrasound images collected from two ultrasound machines across three hospitals from two institutions. The scientific community's enthusiastic participation led to the selection of the top 8 out of 179 entries from 193 registrants in the initial phase to proceed to the competition's second stage. These algorithms have elevated the state-of-the-art in automatic PSFHS from intrapartum ultrasound images. A thorough analysis of the results pinpointed ongoing challenges in the field and outlined recommendations for future work. The top solutions and the complete dataset remain publicly available, fostering further advancements in automatic segmentation and biometry for intrapartum ultrasound imaging.
Generative Diffusion Model Bootstraps Zero-shot Classification of Fetal Ultrasound Images In Underrepresented African Populations
Jul 29, 2024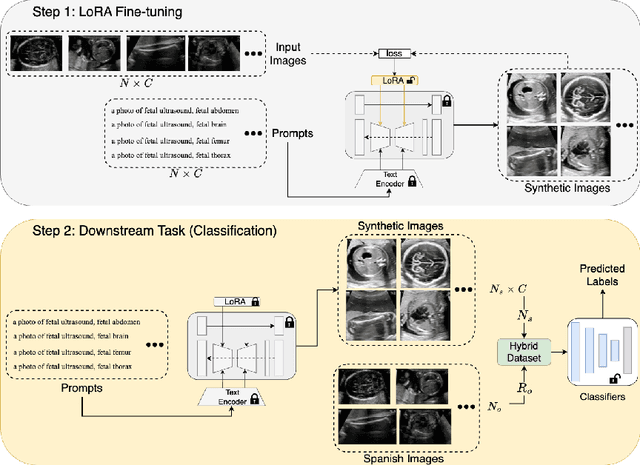
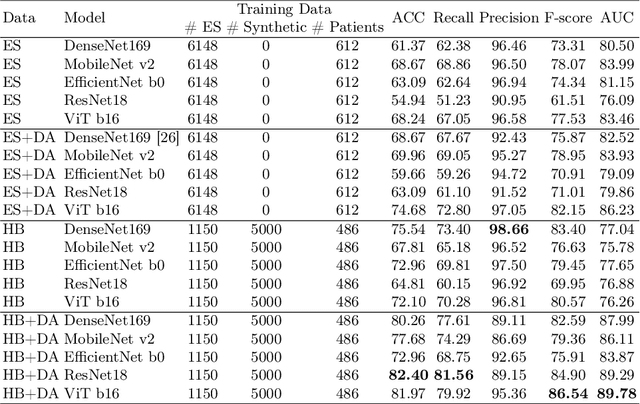
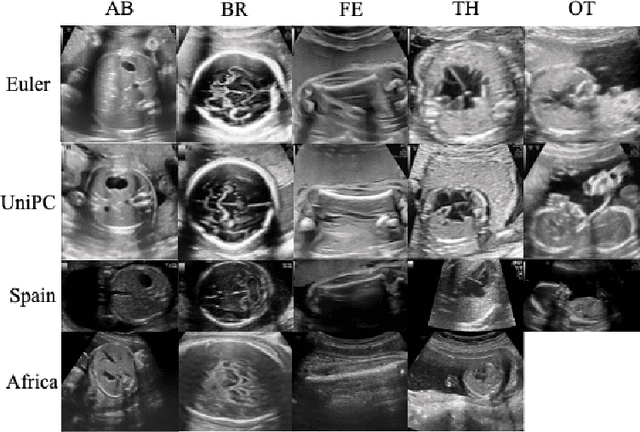
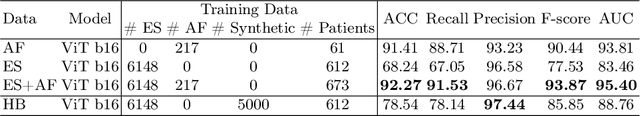
Abstract:Developing robust deep learning models for fetal ultrasound image analysis requires comprehensive, high-quality datasets to effectively learn informative data representations within the domain. However, the scarcity of labelled ultrasound images poses substantial challenges, especially in low-resource settings. To tackle this challenge, we leverage synthetic data to enhance the generalizability of deep learning models. This study proposes a diffusion-based method, Fetal Ultrasound LoRA (FU-LoRA), which involves fine-tuning latent diffusion models using the LoRA technique to generate synthetic fetal ultrasound images. These synthetic images are integrated into a hybrid dataset that combines real-world and synthetic images to improve the performance of zero-shot classifiers in low-resource settings. Our experimental results on fetal ultrasound images from African cohorts demonstrate that FU-LoRA outperforms the baseline method by a 13.73% increase in zero-shot classification accuracy. Furthermore, FU-LoRA achieves the highest accuracy of 82.40%, the highest F-score of 86.54%, and the highest AUC of 89.78%. It demonstrates that the FU-LoRA method is effective in the zero-shot classification of fetal ultrasound images in low-resource settings. Our code and data are publicly accessible on https://github.com/13204942/FU-LoRA.
Segmenting Fetal Head with Efficient Fine-tuning Strategies in Low-resource Settings: an empirical study with U-Net
Jul 29, 2024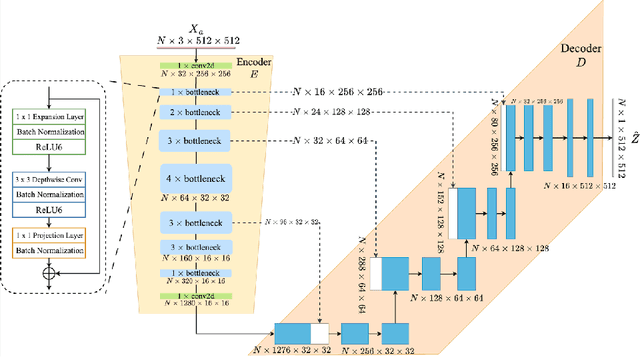
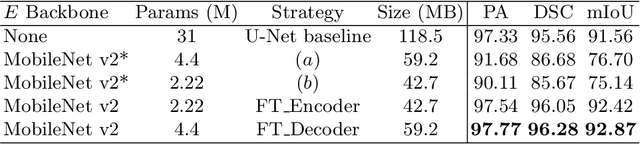
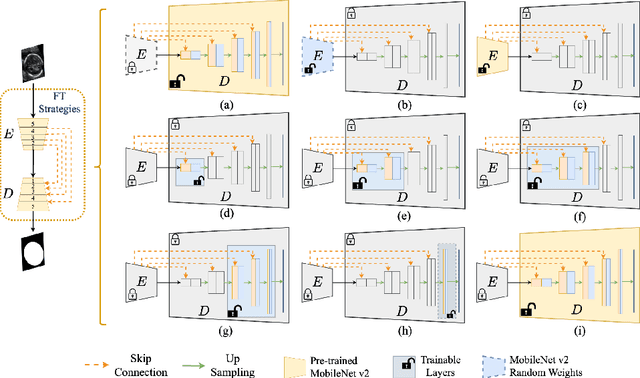
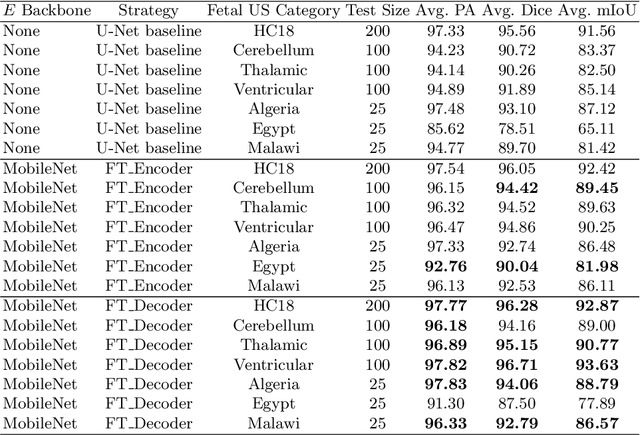
Abstract:Accurate measurement of fetal head circumference is crucial for estimating fetal growth during routine prenatal screening. Prior to measurement, it is necessary to accurately identify and segment the region of interest, specifically the fetal head, in ultrasound images. Recent advancements in deep learning techniques have shown significant progress in segmenting the fetal head using encoder-decoder models. Among these models, U-Net has become a standard approach for accurate segmentation. However, training an encoder-decoder model can be a time-consuming process that demands substantial computational resources. Moreover, fine-tuning these models is particularly challenging when there is a limited amount of data available. There are still no "best-practice" guidelines for optimal fine-tuning of U-net for fetal ultrasound image segmentation. This work summarizes existing fine-tuning strategies with various backbone architectures, model components, and fine-tuning strategies across ultrasound data from Netherlands, Spain, Malawi, Egypt and Algeria. Our study shows that (1) fine-tuning U-Net leads to better performance than training from scratch, (2) fine-tuning strategies in decoder are superior to other strategies, (3) network architecture with less number of parameters can achieve similar or better performance. We also demonstrate the effectiveness of fine-tuning strategies in low-resource settings and further expand our experiments into few-shot learning. Lastly, we publicly released our code and specific fine-tuned weights.
Test-Time Adaptation with SaLIP: A Cascade of SAM and CLIP for Zero shot Medical Image Segmentation
Apr 09, 2024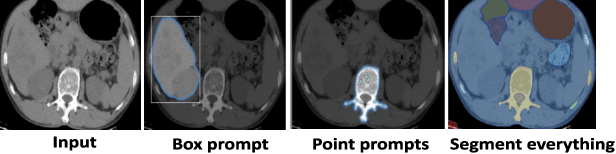
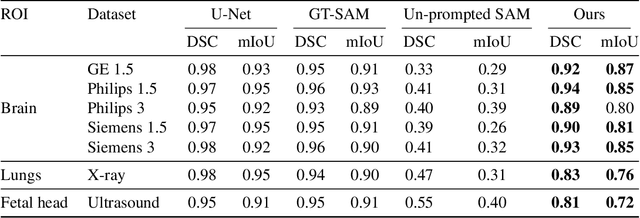
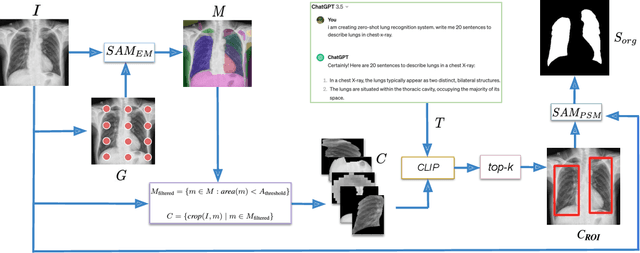

Abstract:The Segment Anything Model (SAM) and CLIP are remarkable vision foundation models (VFMs). SAM, a prompt driven segmentation model, excels in segmentation tasks across diverse domains, while CLIP is renowned for its zero shot recognition capabilities. However, their unified potential has not yet been explored in medical image segmentation. To adapt SAM to medical imaging, existing methods primarily rely on tuning strategies that require extensive data or prior prompts tailored to the specific task, making it particularly challenging when only a limited number of data samples are available. This work presents an in depth exploration of integrating SAM and CLIP into a unified framework for medical image segmentation. Specifically, we propose a simple unified framework, SaLIP, for organ segmentation. Initially, SAM is used for part based segmentation within the image, followed by CLIP to retrieve the mask corresponding to the region of interest (ROI) from the pool of SAM generated masks. Finally, SAM is prompted by the retrieved ROI to segment a specific organ. Thus, SaLIP is training and fine tuning free and does not rely on domain expertise or labeled data for prompt engineering. Our method shows substantial enhancements in zero shot segmentation, showcasing notable improvements in DICE scores across diverse segmentation tasks like brain (63.46%), lung (50.11%), and fetal head (30.82%), when compared to un prompted SAM. Code and text prompts will be available online.
MiTU-Net: A fine-tuned U-Net with SegFormer backbone for segmenting pubic symphysis-fetal head
Jan 27, 2024Abstract:Ultrasound measurements have been examined as potential tools for predicting the likelihood of successful vaginal delivery. The angle of progression (AoP) is a measurable parameter that can be obtained during the initial stage of labor. The AoP is defined as the angle between a straight line along the longitudinal axis of the pubic symphysis (PS) and a line from the inferior edge of the PS to the leading edge of the fetal head (FH). However, the process of measuring AoP on ultrasound images is time consuming and prone to errors. To address this challenge, we propose the Mix Transformer U-Net (MiTU-Net) network, for automatic fetal head-pubic symphysis segmentation and AoP measurement. The MiTU-Net model is based on an encoder-decoder framework, utilizing a pre-trained efficient transformer to enhance feature representation. Within the efficient transformer encoder, the model significantly reduces the trainable parameters of the encoder-decoder model. The effectiveness of the proposed method is demonstrated through experiments conducted on a recent transperineal ultrasound dataset. Our model achieves competitive performance, ranking 5th compared to existing approaches. The MiTU-Net presents an efficient method for automatic segmentation and AoP measurement, reducing errors and assisting sonographers in clinical practice. Reproducibility: Framework implementation and models available on https://github.com/13204942/MiTU-Net.
Lightweight Framework for Automated Kidney Stone Detection using coronal CT images
Nov 24, 2023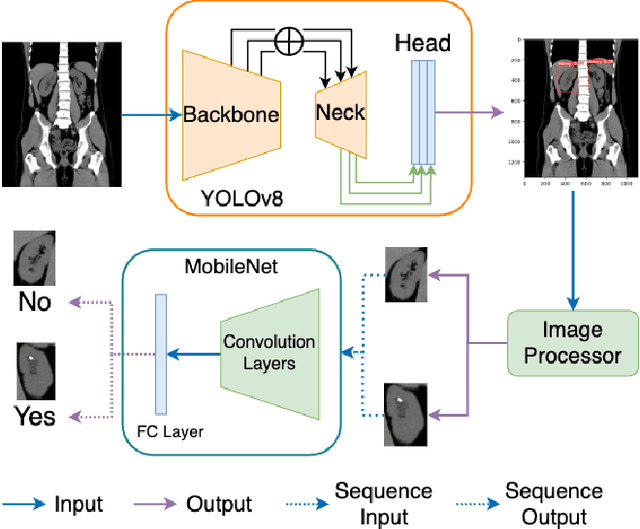


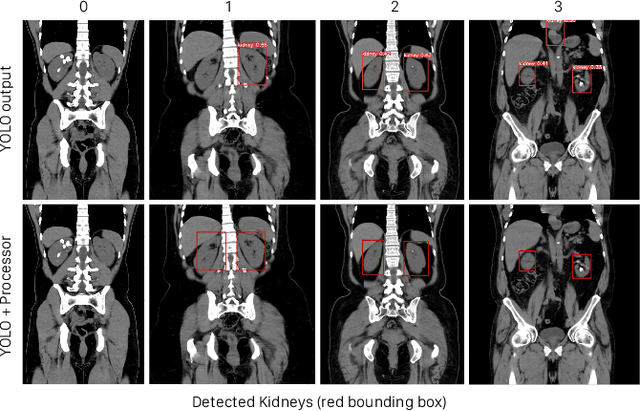
Abstract:Kidney stone disease results in millions of annual visits to emergency departments in the United States. Computed tomography (CT) scans serve as the standard imaging modality for efficient detection of kidney stones. Various approaches utilizing convolutional neural networks (CNNs) have been proposed to implement automatic diagnosis of kidney stones. However, there is a growing interest in employing fast and efficient CNNs on edge devices in clinical practice. In this paper, we propose a lightweight fusion framework for kidney detection and kidney stone diagnosis on coronal CT images. In our design, we aim to minimize the computational costs of training and inference while implementing an automated approach. The experimental results indicate that our framework can achieve competitive outcomes using only 8\% of the original training data. These results include an F1 score of 96\% and a False Negative (FN) error rate of 4\%. Additionally, the average detection time per CT image on a CPU is 0.62 seconds. Reproducibility: Framework implementation and models available on GitHub.
Novel Models for Multiple Dependent Heteroskedastic Time Series
Oct 26, 2023Abstract:Functional magnetic resonance imaging or functional MRI (fMRI) is a very popular tool used for differing brain regions by measuring brain activity. It is affected by physiological noise, such as head and brain movement in the scanner from breathing, heart beats, or the subject fidgeting. The purpose of this paper is to propose a novel approach to handling fMRI data for infants with high volatility caused by sudden head movements. Another purpose is to evaluate the volatility modelling performance of multiple dependent fMRI time series data. The models examined in this paper are AR and GARCH and the modelling performance is evaluated by several statistical performance measures. The conclusions of this paper are that multiple dependent fMRI series data can be fitted with AR + GARCH model if the multiple fMRI data have many sudden head movements. The GARCH model can capture the shared volatility clustering caused by head movements across brain regions. However, the multiple fMRI data without many head movements have fitted AR + GARCH model with different performance. The conclusions are supported by statistical tests and measures. This paper highlights the difference between the proposed approach from traditional approaches when estimating model parameters and modelling conditional variances on multiple dependent time series. In the future, the proposed approach can be applied to other research fields, such as financial economics, and signal processing. Code is available at \url{https://github.com/13204942/STAT40710}.
Evaluate Fine-tuning Strategies for Fetal Head Ultrasound Image Segmentation with U-Net
Jul 18, 2023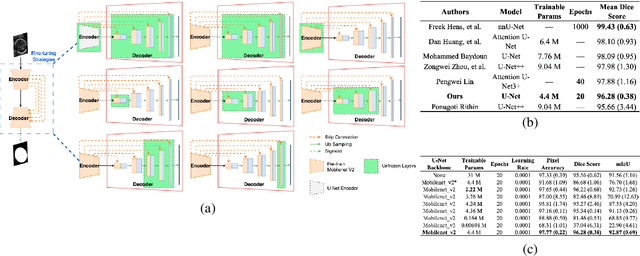
Abstract:Fetal head segmentation is a crucial step in measuring the fetal head circumference (HC) during gestation, an important biometric in obstetrics for monitoring fetal growth. However, manual biometry generation is time-consuming and results in inconsistent accuracy. To address this issue, convolutional neural network (CNN) models have been utilized to improve the efficiency of medical biometry. But training a CNN network from scratch is a challenging task, we proposed a Transfer Learning (TL) method. Our approach involves fine-tuning (FT) a U-Net network with a lightweight MobileNet as the encoder to perform segmentation on a set of fetal head ultrasound (US) images with limited effort. This method addresses the challenges associated with training a CNN network from scratch. It suggests that our proposed FT strategy yields segmentation performance that is comparable when trained with a reduced number of parameters by 85.8%. And our proposed FT strategy outperforms other strategies with smaller trainable parameter sizes below 4.4 million. Thus, we contend that it can serve as a dependable FT approach for reducing the size of models in medical image analysis. Our key findings highlight the importance of the balance between model performance and size in developing Artificial Intelligence (AI) applications by TL methods. Code is available at https://github.com/13204942/FT_Methods_for_Fetal_Head_Segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge