Kathleen M. Curran
Entropy-Guided Agreement-Diversity: A Semi-Supervised Active Learning Framework for Fetal Head Segmentation in Ultrasound
Jan 24, 2026Abstract:Fetal ultrasound (US) data is often limited due to privacy and regulatory restrictions, posing challenges for training deep learning (DL) models. While semi-supervised learning (SSL) is commonly used for fetal US image analysis, existing SSL methods typically rely on random limited selection, which can lead to suboptimal model performance by overfitting to homogeneous labeled data. To address this, we propose a two-stage Active Learning (AL) sampler, Entropy-Guided Agreement-Diversity (EGAD), for fetal head segmentation. Our method first selects the most uncertain samples using predictive entropy, and then refines the final selection using the agreement-diversity score combining cosine similarity and mutual information. Additionally, our SSL framework employs a consistency learning strategy with feature downsampling to further enhance segmentation performance. In experiments, SSL-EGAD achieves an average Dice score of 94.57\% and 96.32\% on two public datasets for fetal head segmentation, using 5\% and 10\% labeled data for training, respectively. Our method outperforms current SSL models and showcases consistent robustness across diverse pregnancy stage data. The code is available on \href{https://github.com/13204942/Semi-supervised-EGAD}{GitHub}.
Parameter-Free Bio-Inspired Channel Attention for Enhanced Cardiac MRI Reconstruction
May 29, 2025Abstract:Attention is a fundamental component of the human visual recognition system. The inclusion of attention in a convolutional neural network amplifies relevant visual features and suppresses the less important ones. Integrating attention mechanisms into convolutional neural networks enhances model performance and interpretability. Spatial and channel attention mechanisms have shown significant advantages across many downstream tasks in medical imaging. While existing attention modules have proven to be effective, their design often lacks a robust theoretical underpinning. In this study, we address this gap by proposing a non-linear attention architecture for cardiac MRI reconstruction and hypothesize that insights from ecological principles can guide the development of effective and efficient attention mechanisms. Specifically, we investigate a non-linear ecological difference equation that describes single-species population growth to devise a parameter-free attention module surpassing current state-of-the-art parameter-free methods.
Segmenting Fetal Head with Efficient Fine-tuning Strategies in Low-resource Settings: an empirical study with U-Net
Jul 29, 2024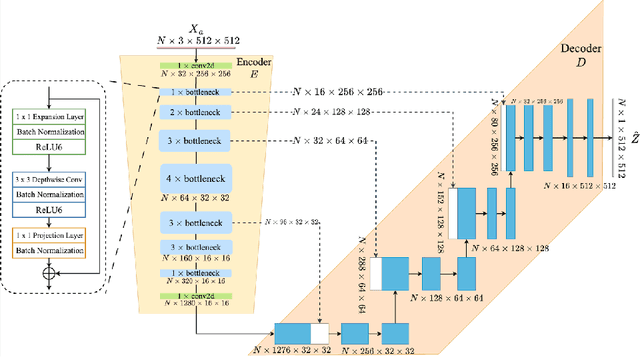
Abstract:Accurate measurement of fetal head circumference is crucial for estimating fetal growth during routine prenatal screening. Prior to measurement, it is necessary to accurately identify and segment the region of interest, specifically the fetal head, in ultrasound images. Recent advancements in deep learning techniques have shown significant progress in segmenting the fetal head using encoder-decoder models. Among these models, U-Net has become a standard approach for accurate segmentation. However, training an encoder-decoder model can be a time-consuming process that demands substantial computational resources. Moreover, fine-tuning these models is particularly challenging when there is a limited amount of data available. There are still no "best-practice" guidelines for optimal fine-tuning of U-net for fetal ultrasound image segmentation. This work summarizes existing fine-tuning strategies with various backbone architectures, model components, and fine-tuning strategies across ultrasound data from Netherlands, Spain, Malawi, Egypt and Algeria. Our study shows that (1) fine-tuning U-Net leads to better performance than training from scratch, (2) fine-tuning strategies in decoder are superior to other strategies, (3) network architecture with less number of parameters can achieve similar or better performance. We also demonstrate the effectiveness of fine-tuning strategies in low-resource settings and further expand our experiments into few-shot learning. Lastly, we publicly released our code and specific fine-tuned weights.
Generative Diffusion Model Bootstraps Zero-shot Classification of Fetal Ultrasound Images In Underrepresented African Populations
Jul 29, 2024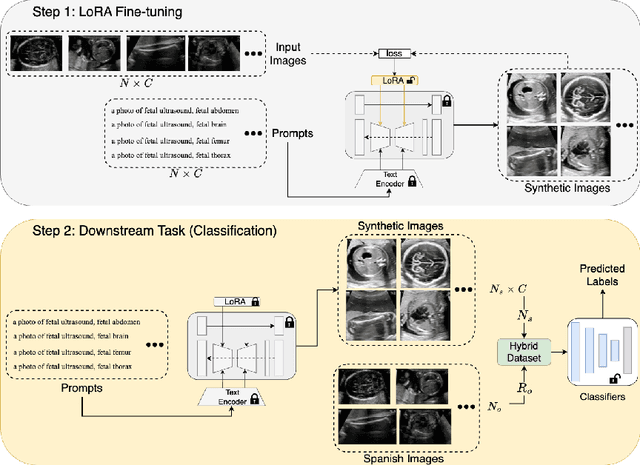
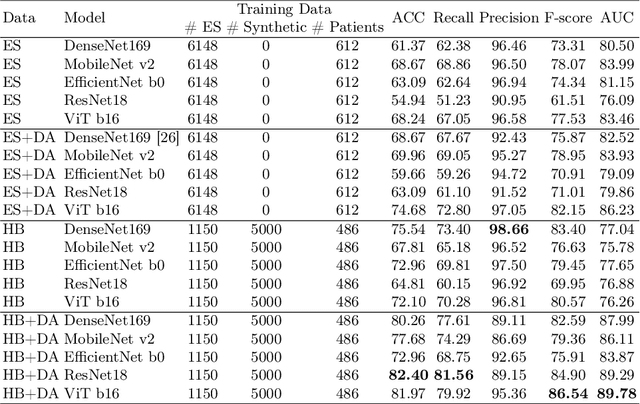
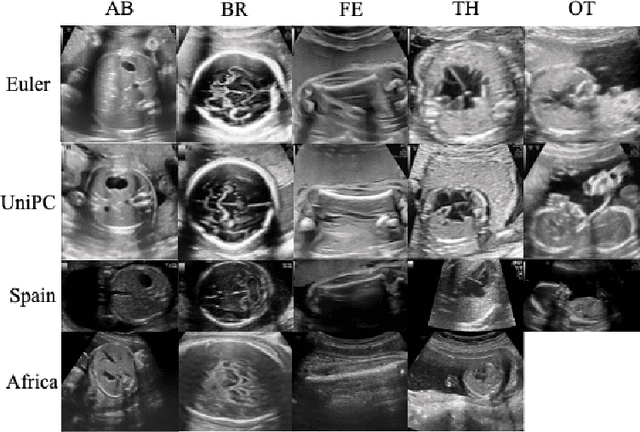
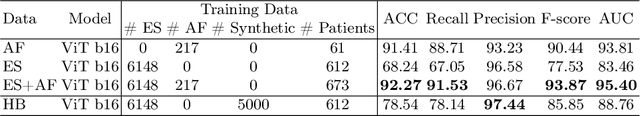
Abstract:Developing robust deep learning models for fetal ultrasound image analysis requires comprehensive, high-quality datasets to effectively learn informative data representations within the domain. However, the scarcity of labelled ultrasound images poses substantial challenges, especially in low-resource settings. To tackle this challenge, we leverage synthetic data to enhance the generalizability of deep learning models. This study proposes a diffusion-based method, Fetal Ultrasound LoRA (FU-LoRA), which involves fine-tuning latent diffusion models using the LoRA technique to generate synthetic fetal ultrasound images. These synthetic images are integrated into a hybrid dataset that combines real-world and synthetic images to improve the performance of zero-shot classifiers in low-resource settings. Our experimental results on fetal ultrasound images from African cohorts demonstrate that FU-LoRA outperforms the baseline method by a 13.73% increase in zero-shot classification accuracy. Furthermore, FU-LoRA achieves the highest accuracy of 82.40%, the highest F-score of 86.54%, and the highest AUC of 89.78%. It demonstrates that the FU-LoRA method is effective in the zero-shot classification of fetal ultrasound images in low-resource settings. Our code and data are publicly accessible on https://github.com/13204942/FU-LoRA.
An AI System for Continuous Knee Osteoarthritis Severity Grading Using Self-Supervised Anomaly Detection with Limited Data
Jul 16, 2024Abstract:The diagnostic accuracy and subjectivity of existing Knee Osteoarthritis (OA) ordinal grading systems has been a subject of on-going debate and concern. Existing automated solutions are trained to emulate these imperfect systems, whilst also being reliant on large annotated databases for fully-supervised training. This work proposes a three stage approach for automated continuous grading of knee OA that is built upon the principles of Anomaly Detection (AD); learning a robust representation of healthy knee X-rays and grading disease severity based on its distance to the centre of normality. In the first stage, SS-FewSOME is proposed, a self-supervised AD technique that learns the 'normal' representation, requiring only examples of healthy subjects and <3% of the labels that existing methods require. In the second stage, this model is used to pseudo label a subset of unlabelled data as 'normal' or 'anomalous', followed by denoising of pseudo labels with CLIP. The final stage involves retraining on labelled and pseudo labelled data using the proposed Dual Centre Representation Learning (DCRL) which learns the centres of two representation spaces; normal and anomalous. Disease severity is then graded based on the distance to the learned centres. The proposed methodology outperforms existing techniques by margins of up to 24% in terms of OA detection and the disease severity scores correlate with the Kellgren-Lawrence grading system at the same level as human expert performance. Code available at https://github.com/niamhbelton/SS-FewSOME_Disease_Severity_Knee_Osteoarthritis.
Accelerating Cardiac MRI Reconstruction with CMRatt: An Attention-Driven Approach
Apr 10, 2024

Abstract:Cine cardiac magnetic resonance (CMR) imaging is recognised as the benchmark modality for the comprehensive assessment of cardiac function. Nevertheless, the acquisition process of cine CMR is considered as an impediment due to its prolonged scanning time. One commonly used strategy to expedite the acquisition process is through k-space undersampling, though it comes with a drawback of introducing aliasing effects in the reconstructed image. Lately, deep learning-based methods have shown remarkable results over traditional approaches in rapidly achieving precise CMR reconstructed images. This study aims to explore the untapped potential of attention mechanisms incorporated with a deep learning model within the context of the CMR reconstruction problem. We are motivated by the fact that attention has proven beneficial in downstream tasks such as image classification and segmentation, but has not been systematically analysed in the context of CMR reconstruction. Our primary goal is to identify the strengths and potential limitations of attention algorithms when integrated with a convolutional backbone model such as a U-Net. To achieve this, we benchmark different state-of-the-art spatial and channel attention mechanisms on the CMRxRecon dataset and quantitatively evaluate the quality of reconstruction using objective metrics. Furthermore, inspired by the best performing attention mechanism, we propose a new, simple yet effective, attention pipeline specifically optimised for the task of cardiac image reconstruction that outperforms other state-of-the-art attention methods. The layer and model code will be made publicly available.
Lightweight Framework for Automated Kidney Stone Detection using coronal CT images
Nov 24, 2023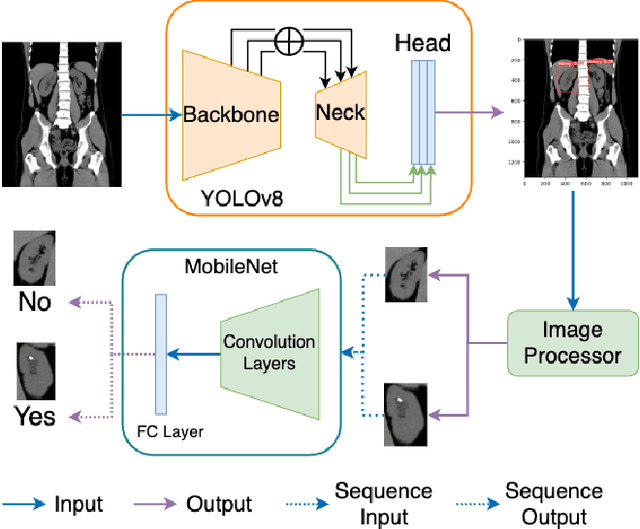


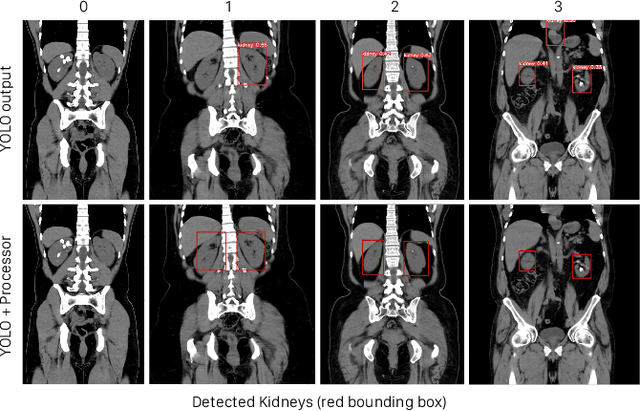
Abstract:Kidney stone disease results in millions of annual visits to emergency departments in the United States. Computed tomography (CT) scans serve as the standard imaging modality for efficient detection of kidney stones. Various approaches utilizing convolutional neural networks (CNNs) have been proposed to implement automatic diagnosis of kidney stones. However, there is a growing interest in employing fast and efficient CNNs on edge devices in clinical practice. In this paper, we propose a lightweight fusion framework for kidney detection and kidney stone diagnosis on coronal CT images. In our design, we aim to minimize the computational costs of training and inference while implementing an automated approach. The experimental results indicate that our framework can achieve competitive outcomes using only 8\% of the original training data. These results include an F1 score of 96\% and a False Negative (FN) error rate of 4\%. Additionally, the average detection time per CT image on a CPU is 0.62 seconds. Reproducibility: Framework implementation and models available on GitHub.
Distance-Aware eXplanation Based Learning
Sep 11, 2023



Abstract:eXplanation Based Learning (XBL) is an interactive learning approach that provides a transparent method of training deep learning models by interacting with their explanations. XBL augments loss functions to penalize a model based on deviation of its explanations from user annotation of image features. The literature on XBL mostly depends on the intersection of visual model explanations and image feature annotations. We present a method to add a distance-aware explanation loss to categorical losses that trains a learner to focus on important regions of a training dataset. Distance is an appropriate approach for calculating explanation loss since visual model explanations such as Gradient-weighted Class Activation Mapping (Grad-CAMs) are not strictly bounded as annotations and their intersections may not provide complete information on the deviation of a model's focus from relevant image regions. In addition to assessing our model using existing metrics, we propose an interpretability metric for evaluating visual feature-attribution based model explanations that is more informative of the model's performance than existing metrics. We demonstrate performance of our proposed method on three image classification tasks.
Unlearning Spurious Correlations in Chest X-ray Classification
Aug 03, 2023Abstract:Medical image classification models are frequently trained using training datasets derived from multiple data sources. While leveraging multiple data sources is crucial for achieving model generalization, it is important to acknowledge that the diverse nature of these sources inherently introduces unintended confounders and other challenges that can impact both model accuracy and transparency. A notable confounding factor in medical image classification, particularly in musculoskeletal image classification, is skeletal maturation-induced bone growth observed during adolescence. We train a deep learning model using a Covid-19 chest X-ray dataset and we showcase how this dataset can lead to spurious correlations due to unintended confounding regions. eXplanation Based Learning (XBL) is a deep learning approach that goes beyond interpretability by utilizing model explanations to interactively unlearn spurious correlations. This is achieved by integrating interactive user feedback, specifically feature annotations. In our study, we employed two non-demanding manual feedback mechanisms to implement an XBL-based approach for effectively eliminating these spurious correlations. Our results underscore the promising potential of XBL in constructing robust models even in the presence of confounding factors.
Evaluate Fine-tuning Strategies for Fetal Head Ultrasound Image Segmentation with U-Net
Jul 18, 2023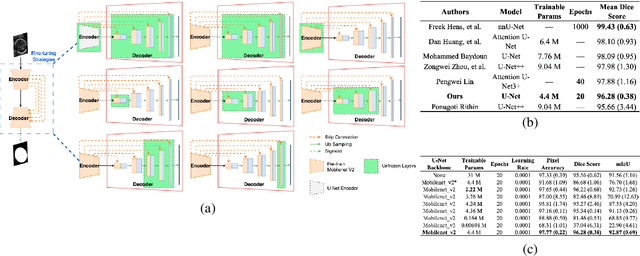
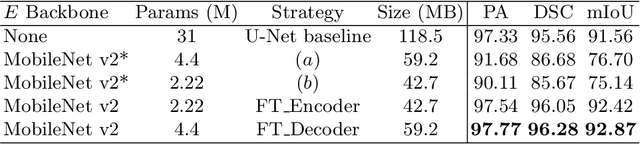
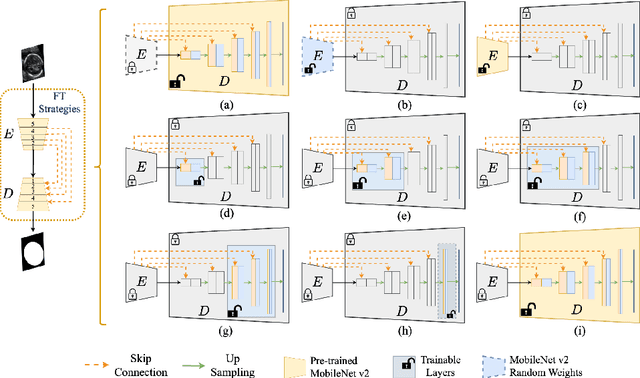
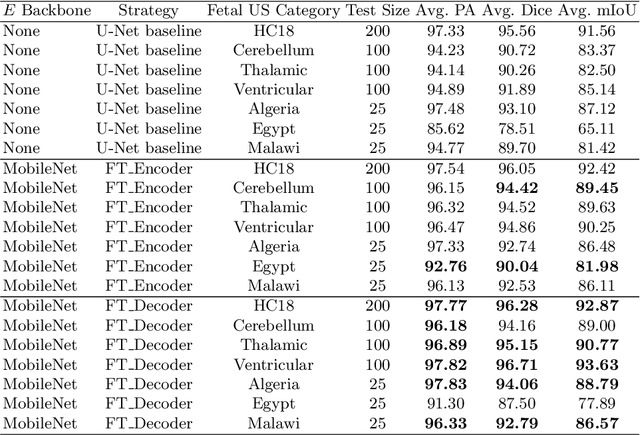
Abstract:Fetal head segmentation is a crucial step in measuring the fetal head circumference (HC) during gestation, an important biometric in obstetrics for monitoring fetal growth. However, manual biometry generation is time-consuming and results in inconsistent accuracy. To address this issue, convolutional neural network (CNN) models have been utilized to improve the efficiency of medical biometry. But training a CNN network from scratch is a challenging task, we proposed a Transfer Learning (TL) method. Our approach involves fine-tuning (FT) a U-Net network with a lightweight MobileNet as the encoder to perform segmentation on a set of fetal head ultrasound (US) images with limited effort. This method addresses the challenges associated with training a CNN network from scratch. It suggests that our proposed FT strategy yields segmentation performance that is comparable when trained with a reduced number of parameters by 85.8%. And our proposed FT strategy outperforms other strategies with smaller trainable parameter sizes below 4.4 million. Thus, we contend that it can serve as a dependable FT approach for reducing the size of models in medical image analysis. Our key findings highlight the importance of the balance between model performance and size in developing Artificial Intelligence (AI) applications by TL methods. Code is available at https://github.com/13204942/FT_Methods_for_Fetal_Head_Segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge