Eric Arazo
Uncertainty Makes It Stable: Curiosity-Driven Quantized Mixture-of-Experts
Nov 19, 2025Abstract:Deploying deep neural networks on resource-constrained devices faces two critical challenges: maintaining accuracy under aggressive quantization while ensuring predictable inference latency. We present a curiosity-driven quantized Mixture-of-Experts framework that addresses both through Bayesian epistemic uncertainty-based routing across heterogeneous experts (BitNet ternary, 1-16 bit BitLinear, post-training quantization). Evaluated on audio classification benchmarks (ESC-50, Quinn, UrbanSound8K), our 4-bit quantization maintains 99.9 percent of 16-bit accuracy (0.858 vs 0.859 F1) with 4x compression and 41 percent energy savings versus 8-bit. Crucially, curiosity-driven routing reduces MoE latency variance by 82 percent (p = 0.008, Levene's test) from 230 ms to 29 ms standard deviation, enabling stable inference for battery-constrained devices. Statistical analysis confirms 4-bit/8-bit achieve practical equivalence with full precision (p > 0.05), while MoE architectures introduce 11 percent latency overhead (p < 0.001) without accuracy gains. At scale, deployment emissions dominate training by 10000x for models serving more than 1,000 inferences, making inference efficiency critical. Our information-theoretic routing demonstrates that adaptive quantization yields accurate (0.858 F1, 1.2M params), energy-efficient (3.87 F1/mJ), and predictable edge models, with simple 4-bit quantized architectures outperforming complex MoE for most deployments.
An accurate detection is not all you need to combat label noise in web-noisy datasets
Jul 08, 2024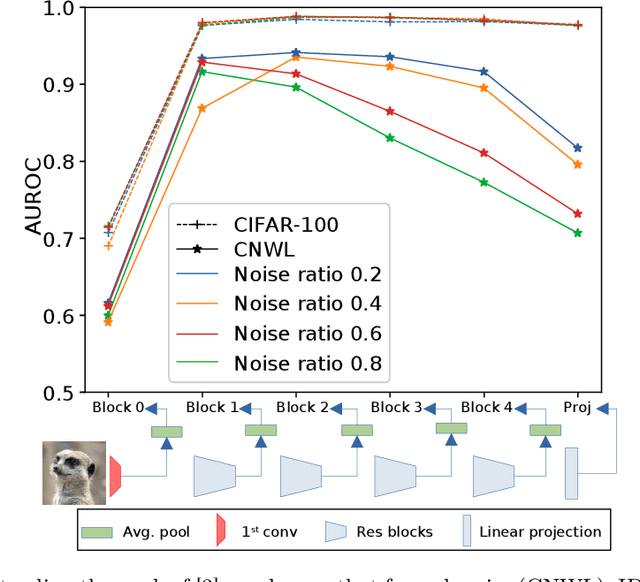
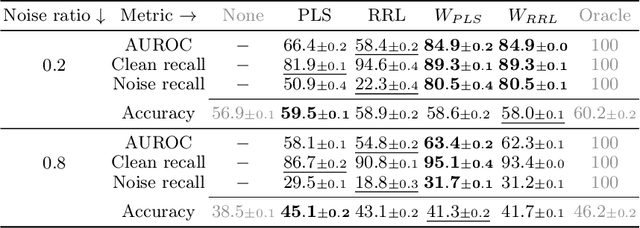

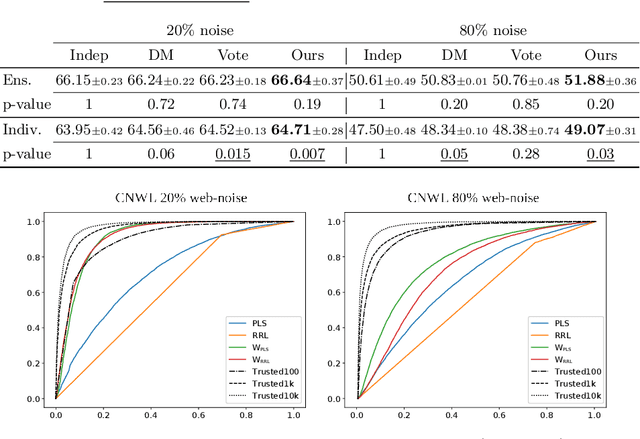
Abstract:Training a classifier on web-crawled data demands learning algorithms that are robust to annotation errors and irrelevant examples. This paper builds upon the recent empirical observation that applying unsupervised contrastive learning to noisy, web-crawled datasets yields a feature representation under which the in-distribution (ID) and out-of-distribution (OOD) samples are linearly separable. We show that direct estimation of the separating hyperplane can indeed offer an accurate detection of OOD samples, and yet, surprisingly, this detection does not translate into gains in classification accuracy. Digging deeper into this phenomenon, we discover that the near-perfect detection misses a type of clean examples that are valuable for supervised learning. These examples often represent visually simple images, which are relatively easy to identify as clean examples using standard loss- or distance-based methods despite being poorly separated from the OOD distribution using unsupervised learning. Because we further observe a low correlation with SOTA metrics, this urges us to propose a hybrid solution that alternates between noise detection using linear separation and a state-of-the-art (SOTA) small-loss approach. When combined with the SOTA algorithm PLS, we substantially improve SOTA results for real-world image classification in the presence of web noise github.com/PaulAlbert31/LSA
Test-Time Adaptation with SaLIP: A Cascade of SAM and CLIP for Zero shot Medical Image Segmentation
Apr 09, 2024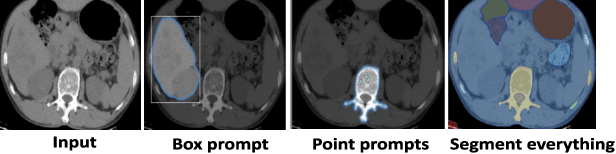
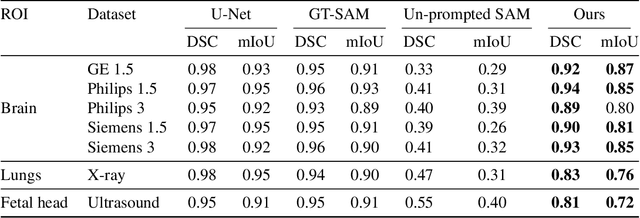
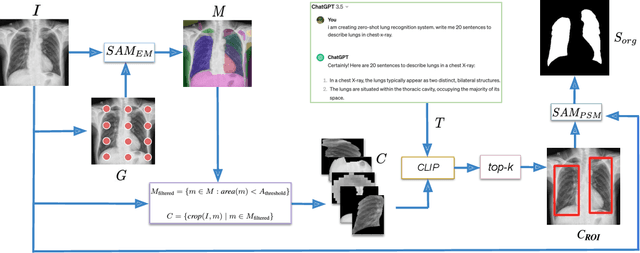

Abstract:The Segment Anything Model (SAM) and CLIP are remarkable vision foundation models (VFMs). SAM, a prompt driven segmentation model, excels in segmentation tasks across diverse domains, while CLIP is renowned for its zero shot recognition capabilities. However, their unified potential has not yet been explored in medical image segmentation. To adapt SAM to medical imaging, existing methods primarily rely on tuning strategies that require extensive data or prior prompts tailored to the specific task, making it particularly challenging when only a limited number of data samples are available. This work presents an in depth exploration of integrating SAM and CLIP into a unified framework for medical image segmentation. Specifically, we propose a simple unified framework, SaLIP, for organ segmentation. Initially, SAM is used for part based segmentation within the image, followed by CLIP to retrieve the mask corresponding to the region of interest (ROI) from the pool of SAM generated masks. Finally, SAM is prompted by the retrieved ROI to segment a specific organ. Thus, SaLIP is training and fine tuning free and does not rely on domain expertise or labeled data for prompt engineering. Our method shows substantial enhancements in zero shot segmentation, showcasing notable improvements in DICE scores across diverse segmentation tasks like brain (63.46%), lung (50.11%), and fetal head (30.82%), when compared to un prompted SAM. Code and text prompts will be available online.
ConvLoRA and AdaBN based Domain Adaptation via Self-Training
Feb 07, 2024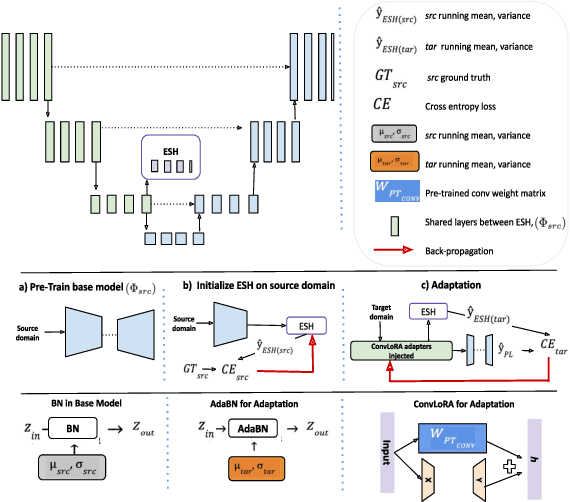
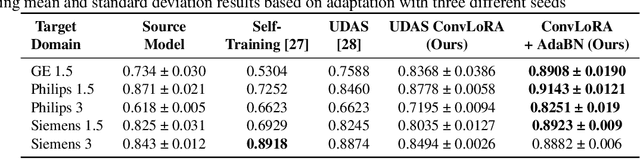
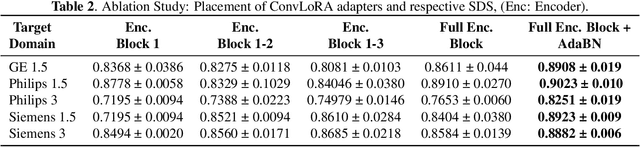
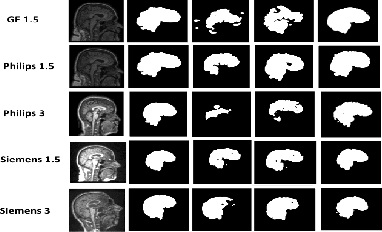
Abstract:Existing domain adaptation (DA) methods often involve pre-training on the source domain and fine-tuning on the target domain. For multi-target domain adaptation, having a dedicated/separate fine-tuned network for each target domain, that retain all the pre-trained model parameters, is prohibitively expensive. To address this limitation, we propose Convolutional Low-Rank Adaptation (ConvLoRA). ConvLoRA freezes pre-trained model weights, adds trainable low-rank decomposition matrices to convolutional layers, and backpropagates the gradient through these matrices thus greatly reducing the number of trainable parameters. To further boost adaptation, we utilize Adaptive Batch Normalization (AdaBN) which computes target-specific running statistics and use it along with ConvLoRA. Our method has fewer trainable parameters and performs better or on-par with large independent fine-tuned networks (with less than 0.9% trainable parameters of the total base model) when tested on the segmentation of Calgary-Campinas dataset containing brain MRI images. Our approach is simple, yet effective and can be applied to any deep learning-based architecture which uses convolutional and batch normalization layers. Code is available at: https://github.com/aleemsidra/ConvLoRA.
Self-Supervised and Semi-Supervised Polyp Segmentation using Synthetic Data
Jul 22, 2023



Abstract:Early detection of colorectal polyps is of utmost importance for their treatment and for colorectal cancer prevention. Computer vision techniques have the potential to aid professionals in the diagnosis stage, where colonoscopies are manually carried out to examine the entirety of the patient's colon. The main challenge in medical imaging is the lack of data, and a further challenge specific to polyp segmentation approaches is the difficulty of manually labeling the available data: the annotation process for segmentation tasks is very time-consuming. While most recent approaches address the data availability challenge with sophisticated techniques to better exploit the available labeled data, few of them explore the self-supervised or semi-supervised paradigm, where the amount of labeling required is greatly reduced. To address both challenges, we leverage synthetic data and propose an end-to-end model for polyp segmentation that integrates real and synthetic data to artificially increase the size of the datasets and aid the training when unlabeled samples are available. Concretely, our model, Pl-CUT-Seg, transforms synthetic images with an image-to-image translation module and combines the resulting images with real images to train a segmentation model, where we use model predictions as pseudo-labels to better leverage unlabeled samples. Additionally, we propose PL-CUT-Seg+, an improved version of the model that incorporates targeted regularization to address the domain gap between real and synthetic images. The models are evaluated on standard benchmarks for polyp segmentation and reach state-of-the-art results in the self- and semi-supervised setups.
Joint one-sided synthetic unpaired image translation and segmentation for colorectal cancer prevention
Jul 20, 2023Abstract:Deep learning has shown excellent performance in analysing medical images. However, datasets are difficult to obtain due privacy issues, standardization problems, and lack of annotations. We address these problems by producing realistic synthetic images using a combination of 3D technologies and generative adversarial networks. We propose CUT-seg, a joint training where a segmentation model and a generative model are jointly trained to produce realistic images while learning to segment polyps. We take advantage of recent one-sided translation models because they use significantly less memory, allowing us to add a segmentation model in the training loop. CUT-seg performs better, is computationally less expensive, and requires less real images than other memory-intensive image translation approaches that require two stage training. Promising results are achieved on five real polyp segmentation datasets using only one real image and zero real annotations. As a part of this study we release Synth-Colon, an entirely synthetic dataset that includes 20000 realistic colon images and additional details about depth and 3D geometry: https://enric1994.github.io/synth-colon
Is your noise correction noisy? PLS: Robustness to label noise with two stage detection
Oct 10, 2022



Abstract:Designing robust algorithms capable of training accurate neural networks on uncurated datasets from the web has been the subject of much research as it reduces the need for time consuming human labor. The focus of many previous research contributions has been on the detection of different types of label noise; however, this paper proposes to improve the correction accuracy of noisy samples once they have been detected. In many state-of-the-art contributions, a two phase approach is adopted where the noisy samples are detected before guessing a corrected pseudo-label in a semi-supervised fashion. The guessed pseudo-labels are then used in the supervised objective without ensuring that the label guess is likely to be correct. This can lead to confirmation bias, which reduces the noise robustness. Here we propose the pseudo-loss, a simple metric that we find to be strongly correlated with pseudo-label correctness on noisy samples. Using the pseudo-loss, we dynamically down weight under-confident pseudo-labels throughout training to avoid confirmation bias and improve the network accuracy. We additionally propose to use a confidence guided contrastive objective that learns robust representation on an interpolated objective between class bound (supervised) for confidently corrected samples and unsupervised representation for under-confident label corrections. Experiments demonstrate the state-of-the-art performance of our Pseudo-Loss Selection (PLS) algorithm on a variety of benchmark datasets including curated data synthetically corrupted with in-distribution and out-of-distribution noise, and two real world web noise datasets. Our experiments are fully reproducible [github coming soon]
Cardiac Segmentation using Transfer Learning under Respiratory Motion Artifacts
Sep 20, 2022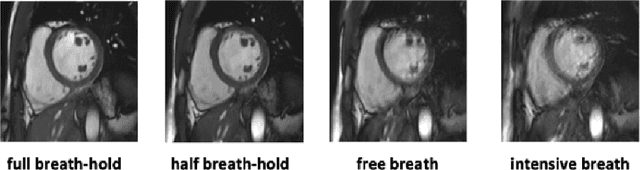
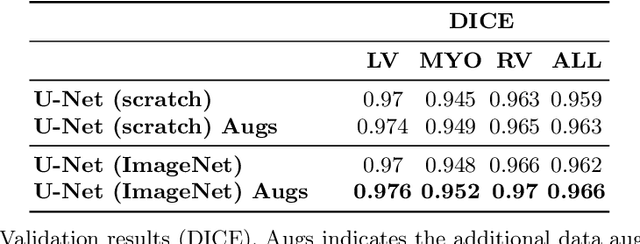
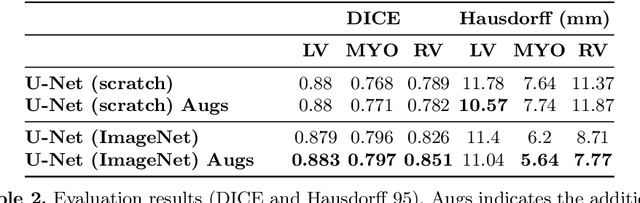
Abstract:Methods that are resilient to artifacts in the cardiac magnetic resonance imaging (MRI) while performing ventricle segmentation, are crucial for ensuring quality in structural and functional analysis of those tissues. While there has been significant efforts on improving the quality of the algorithms, few works have tackled the harm that the artifacts generate in the predictions. In this work, we study fine tuning of pretrained networks to improve the resilience of previous methods to these artifacts. In our proposed method, we adopted the extensive usage of data augmentations that mimic those artifacts. The results significantly improved the baseline segmentations (up to 0.06 Dice score, and 4mm Hausdorff distance improvement).
Embedding contrastive unsupervised features to cluster in- and out-of-distribution noise in corrupted image datasets
Jul 18, 2022
Abstract:Using search engines for web image retrieval is a tempting alternative to manual curation when creating an image dataset, but their main drawback remains the proportion of incorrect (noisy) samples retrieved. These noisy samples have been evidenced by previous works to be a mixture of in-distribution (ID) samples, assigned to the incorrect category but presenting similar visual semantics to other classes in the dataset, and out-of-distribution (OOD) images, which share no semantic correlation with any category from the dataset. The latter are, in practice, the dominant type of noisy images retrieved. To tackle this noise duality, we propose a two stage algorithm starting with a detection step where we use unsupervised contrastive feature learning to represent images in a feature space. We find that the alignment and uniformity principles of contrastive learning allow OOD samples to be linearly separated from ID samples on the unit hypersphere. We then spectrally embed the unsupervised representations using a fixed neighborhood size and apply an outlier sensitive clustering at the class level to detect the clean and OOD clusters as well as ID noisy outliers. We finally train a noise robust neural network that corrects ID noise to the correct category and utilizes OOD samples in a guided contrastive objective, clustering them to improve low-level features. Our algorithm improves the state-of-the-art results on synthetic noise image datasets as well as real-world web-crawled data. Our work is fully reproducible github.com/PaulAlbert31/SNCF.
Segmentation Enhanced Lameness Detection in Dairy Cows from RGB and Depth Video
Jun 09, 2022



Abstract:Cow lameness is a severe condition that affects the life cycle and life quality of dairy cows and results in considerable economic losses. Early lameness detection helps farmers address illnesses early and avoid negative effects caused by the degeneration of cows' condition. We collected a dataset of short clips of cows passing through a hallway exiting a milking station and annotated the degree of lameness of the cows. This paper explores the resulting dataset and provides a detailed description of the data collection process. Additionally, we proposed a lameness detection method that leverages pre-trained neural networks to extract discriminative features from videos and assign a binary score to each cow indicating its condition: "healthy" or "lame." We improve this approach by forcing the model to focus on the structure of the cow, which we achieve by substituting the RGB videos with binary segmentation masks predicted with a trained segmentation model. This work aims to encourage research and provide insights into the applicability of computer vision models for cow lameness detection on farms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge