Kathleen Curran
Towards generating more interpretable counterfactuals via concept vectors: a preliminary study on chest X-rays
Jun 04, 2025Abstract:An essential step in deploying medical imaging models is ensuring alignment with clinical knowledge and interpretability. We focus on mapping clinical concepts into the latent space of generative models to identify Concept Activation Vectors (CAVs). Using a simple reconstruction autoencoder, we link user-defined concepts to image-level features without explicit label training. The extracted concepts are stable across datasets, enabling visual explanations that highlight clinically relevant features. By traversing latent space along concept directions, we produce counterfactuals that exaggerate or reduce specific clinical features. Preliminary results on chest X-rays show promise for large pathologies like cardiomegaly, while smaller pathologies remain challenging due to reconstruction limits. Although not outperforming baselines, this approach offers a path toward interpretable, concept-based explanations aligned with clinical knowledge.
PSFHS Challenge Report: Pubic Symphysis and Fetal Head Segmentation from Intrapartum Ultrasound Images
Sep 17, 2024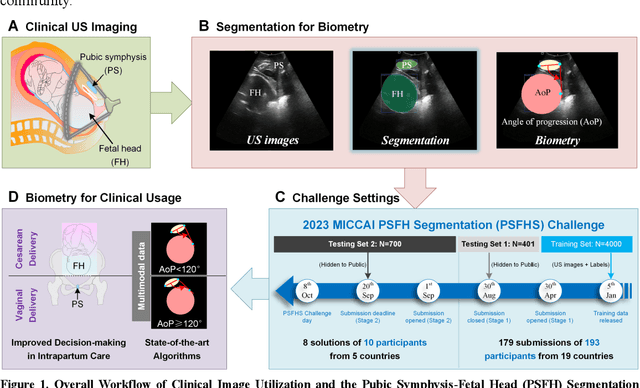
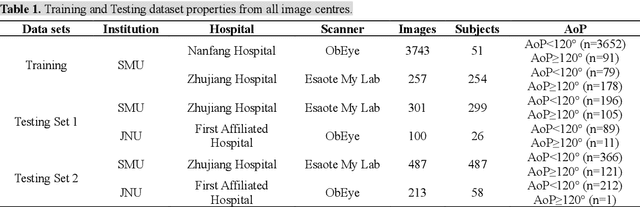
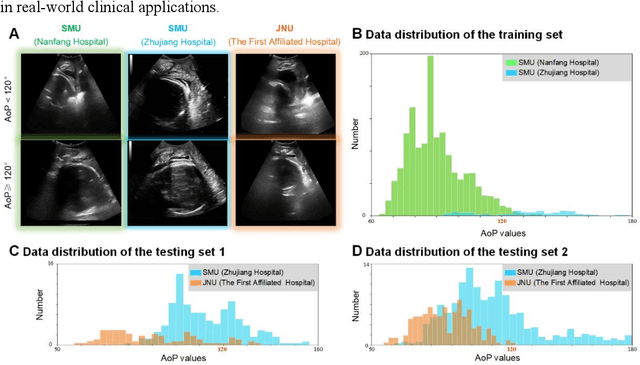

Abstract:Segmentation of the fetal and maternal structures, particularly intrapartum ultrasound imaging as advocated by the International Society of Ultrasound in Obstetrics and Gynecology (ISUOG) for monitoring labor progression, is a crucial first step for quantitative diagnosis and clinical decision-making. This requires specialized analysis by obstetrics professionals, in a task that i) is highly time- and cost-consuming and ii) often yields inconsistent results. The utility of automatic segmentation algorithms for biometry has been proven, though existing results remain suboptimal. To push forward advancements in this area, the Grand Challenge on Pubic Symphysis-Fetal Head Segmentation (PSFHS) was held alongside the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to enhance the development of automatic segmentation algorithms at an international scale, providing the largest dataset to date with 5,101 intrapartum ultrasound images collected from two ultrasound machines across three hospitals from two institutions. The scientific community's enthusiastic participation led to the selection of the top 8 out of 179 entries from 193 registrants in the initial phase to proceed to the competition's second stage. These algorithms have elevated the state-of-the-art in automatic PSFHS from intrapartum ultrasound images. A thorough analysis of the results pinpointed ongoing challenges in the field and outlined recommendations for future work. The top solutions and the complete dataset remain publicly available, fostering further advancements in automatic segmentation and biometry for intrapartum ultrasound imaging.
Test-Time Adaptation with SaLIP: A Cascade of SAM and CLIP for Zero shot Medical Image Segmentation
Apr 09, 2024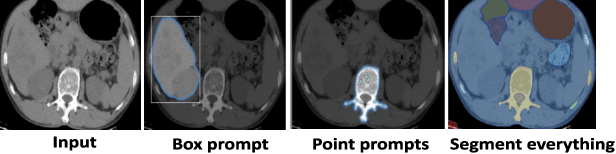
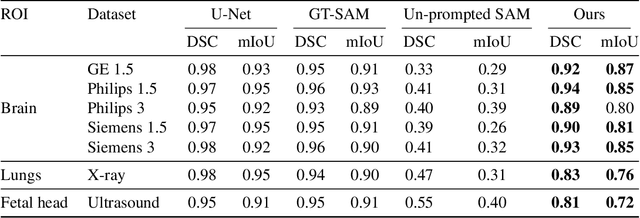
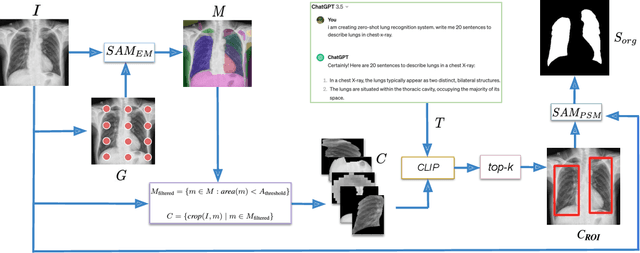

Abstract:The Segment Anything Model (SAM) and CLIP are remarkable vision foundation models (VFMs). SAM, a prompt driven segmentation model, excels in segmentation tasks across diverse domains, while CLIP is renowned for its zero shot recognition capabilities. However, their unified potential has not yet been explored in medical image segmentation. To adapt SAM to medical imaging, existing methods primarily rely on tuning strategies that require extensive data or prior prompts tailored to the specific task, making it particularly challenging when only a limited number of data samples are available. This work presents an in depth exploration of integrating SAM and CLIP into a unified framework for medical image segmentation. Specifically, we propose a simple unified framework, SaLIP, for organ segmentation. Initially, SAM is used for part based segmentation within the image, followed by CLIP to retrieve the mask corresponding to the region of interest (ROI) from the pool of SAM generated masks. Finally, SAM is prompted by the retrieved ROI to segment a specific organ. Thus, SaLIP is training and fine tuning free and does not rely on domain expertise or labeled data for prompt engineering. Our method shows substantial enhancements in zero shot segmentation, showcasing notable improvements in DICE scores across diverse segmentation tasks like brain (63.46%), lung (50.11%), and fetal head (30.82%), when compared to un prompted SAM. Code and text prompts will be available online.
MiTU-Net: A fine-tuned U-Net with SegFormer backbone for segmenting pubic symphysis-fetal head
Jan 27, 2024Abstract:Ultrasound measurements have been examined as potential tools for predicting the likelihood of successful vaginal delivery. The angle of progression (AoP) is a measurable parameter that can be obtained during the initial stage of labor. The AoP is defined as the angle between a straight line along the longitudinal axis of the pubic symphysis (PS) and a line from the inferior edge of the PS to the leading edge of the fetal head (FH). However, the process of measuring AoP on ultrasound images is time consuming and prone to errors. To address this challenge, we propose the Mix Transformer U-Net (MiTU-Net) network, for automatic fetal head-pubic symphysis segmentation and AoP measurement. The MiTU-Net model is based on an encoder-decoder framework, utilizing a pre-trained efficient transformer to enhance feature representation. Within the efficient transformer encoder, the model significantly reduces the trainable parameters of the encoder-decoder model. The effectiveness of the proposed method is demonstrated through experiments conducted on a recent transperineal ultrasound dataset. Our model achieves competitive performance, ranking 5th compared to existing approaches. The MiTU-Net presents an efficient method for automatic segmentation and AoP measurement, reducing errors and assisting sonographers in clinical practice. Reproducibility: Framework implementation and models available on https://github.com/13204942/MiTU-Net.
Interpretability of a Deep Learning Model in the Application of Cardiac MRI Segmentation with an ACDC Challenge Dataset
Mar 15, 2021
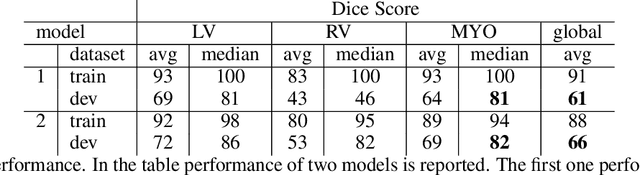

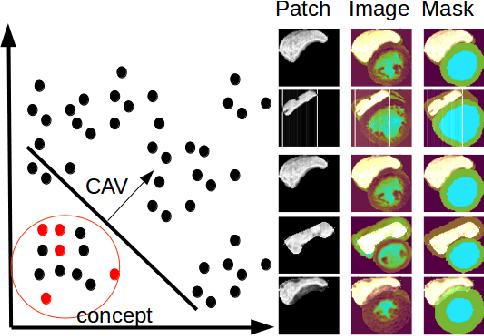
Abstract:Cardiac Magnetic Resonance (CMR) is the most effective tool for the assessment and diagnosis of a heart condition, which malfunction is the world's leading cause of death. Software tools leveraging Artificial Intelligence already enhance radiologists and cardiologists in heart condition assessment but their lack of transparency is a problem. This project investigates if it is possible to discover concepts representative for different cardiac conditions from the deep network trained to segment crdiac structures: Left Ventricle (LV), Right Ventricle (RV) and Myocardium (MYO), using explainability methods that enhances classification system by providing the score-based values of qualitative concepts, along with the key performance metrics. With introduction of a need of explanations in GDPR explainability of AI systems is necessary. This study applies Discovering and Testing with Concept Activation Vectors (D-TCAV), an interpretaibilty method to extract underlying features important for cardiac disease diagnosis from MRI data. The method provides a quantitative notion of concept importance for disease classified. In previous studies, the base method is applied to the classification of cardiac disease and provides clinically meaningful explanations for the predictions of a black-box deep learning classifier. This study applies a method extending TCAV with a Discovering phase (D-TCAV) to cardiac MRI analysis. The advantage of the D-TCAV method over the base method is that it is user-independent. The contribution of this study is a novel application of the explainability method D-TCAV for cardiac MRI anlysis. D-TCAV provides a shorter pre-processing time for clinicians than the base method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge