Kris Sankaran
Department of Statistics, University of Wisconsin - Madison
Data Science Principles for Interpretable and Explainable AI
May 17, 2024Abstract:Society's capacity for algorithmic problem-solving has never been greater. Artificial Intelligence is now applied across more domains than ever, a consequence of powerful abstractions, abundant data, and accessible software. As capabilities have expanded, so have risks, with models often deployed without fully understanding their potential impacts. Interpretable and interactive machine learning aims to make complex models more transparent and controllable, enhancing user agency. This review synthesizes key principles from the growing literature in this field. We first introduce precise vocabulary for discussing interpretability, like the distinction between glass box and explainable algorithms. We then explore connections to classical statistical and design principles, like parsimony and the gulfs of interaction. Basic explainability techniques -- including learned embeddings, integrated gradients, and concept bottlenecks -- are illustrated with a simple case study. We also review criteria for objectively evaluating interpretability approaches. Throughout, we underscore the importance of considering audience goals when designing interactive algorithmic systems. Finally, we outline open challenges and discuss the potential role of data science in addressing them. Code to reproduce all examples can be found at https://go.wisc.edu/3k1ewe.
Spatial Transcriptomics Dimensionality Reduction using Wavelet Bases
May 19, 2022



Abstract:Spatially resolved transcriptomics (ST) measures gene expression along with the spatial coordinates of the measurements. The analysis of ST data involves significant computation complexity. In this work, we propose gene expression dimensionality reduction algorithm that retains spatial structure. We combine the wavelet transformation with matrix factorization to select spatially-varying genes. We extract a low-dimensional representation of these genes. We consider Empirical Bayes setting, imposing regularization through the prior distribution of factor genes. Additionally, We provide visualization of extracted representation genes capturing the global spatial pattern. We illustrate the performance of our methods by spatial structure recovery and gene expression reconstruction in simulation. In real data experiments, our method identifies spatial structure of gene factors and outperforms regular decomposition regarding reconstruction error. We found the connection between the fluctuation of gene patterns and wavelet technique, providing smoother visualization. We develop the package and share the workflow generating reproducible quantitative results and gene visualization. The package is available at https://github.com/OliverXUZY/waveST.
Sampling Strategy for Fine-Tuning Segmentation Models to Crisis Area under Scarcity of Data
Feb 09, 2022Abstract:The use of remote sensing in humanitarian crisis response missions is well-established and has proven relevant repeatedly. One of the problems is obtaining gold annotations as it is costly and time consuming which makes it almost impossible to fine-tune models to new regions affected by the crisis. Where time is critical, resources are limited and environment is constantly changing, models has to evolve and provide flexible ways to adapt to a new situation. The question that we want to answer is if prioritization of samples provide better results in fine-tuning vs other classical sampling methods under annotated data scarcity? We propose a method to guide data collection during fine-tuning, based on estimated model and sample properties, like predicted IOU score. We propose two formulas for calculating sample priority. Our approach blends techniques from interpretability, representation learning and active learning. We have applied our method to a deep learning model for semantic segmentation, U-Net, in a remote sensing application of building detection - one of the core use cases of remote sensing in humanitarian applications. Preliminary results shows utility in prioritization of samples for tuning semantic segmentation models under scarcity of data condition.
Discovering Concepts in Learned Representations using Statistical Inference and Interactive Visualization
Feb 09, 2022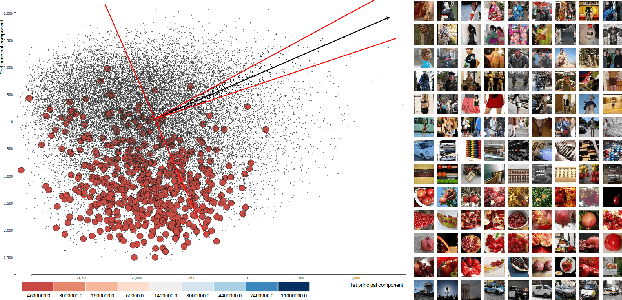
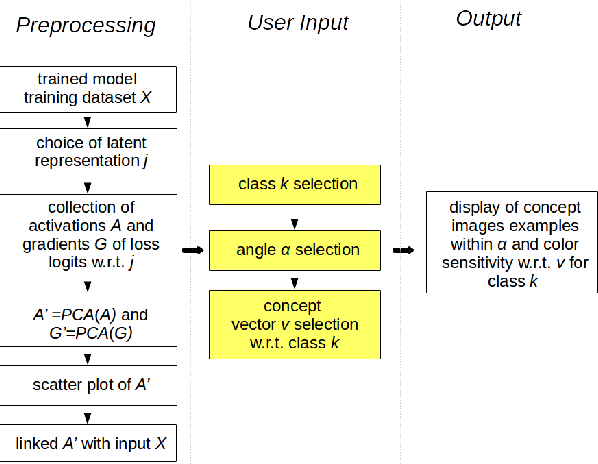
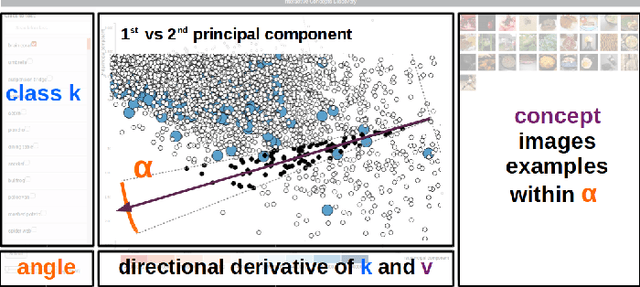
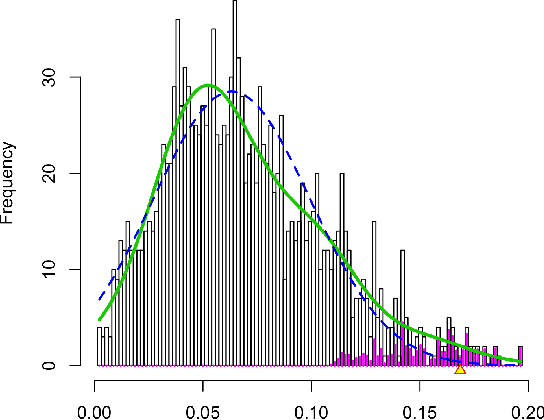
Abstract:Concept discovery is one of the open problems in the interpretability literature that is important for bridging the gap between non-deep learning experts and model end-users. Among current formulations, concepts defines them by as a direction in a learned representation space. This definition makes it possible to evaluate whether a particular concept significantly influences classification decisions for classes of interest. However, finding relevant concepts is tedious, as representation spaces are high-dimensional and hard to navigate. Current approaches include hand-crafting concept datasets and then converting them to latent space directions; alternatively, the process can be automated by clustering the latent space. In this study, we offer another two approaches to guide user discovery of meaningful concepts, one based on multiple hypothesis testing, and another on interactive visualization. We explore the potential value and limitations of these approaches through simulation experiments and an demo visual interface to real data. Overall, we find that these techniques offer a promising strategy for discovering relevant concepts in settings where users do not have predefined descriptions of them, but without completely automating the process.
Source data selection for out-of-domain generalization
Feb 04, 2022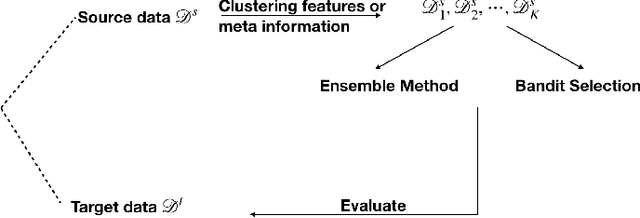
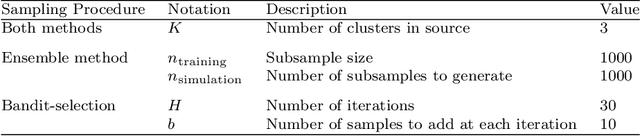
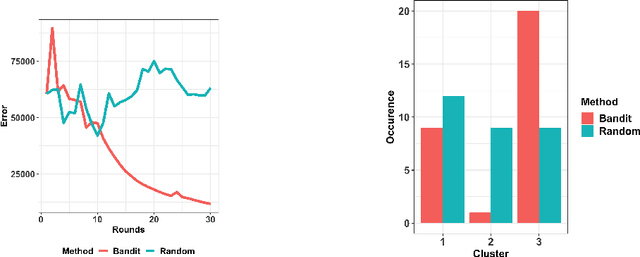
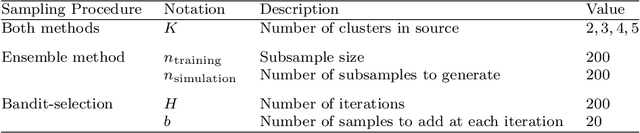
Abstract:Models that perform out-of-domain generalization borrow knowledge from heterogeneous source data and apply it to a related but distinct target task. Transfer learning has proven effective for accomplishing this generalization in many applications. However, poor selection of a source dataset can lead to poor performance on the target, a phenomenon called negative transfer. In order to take full advantage of available source data, this work studies source data selection with respect to a target task. We propose two source selection methods that are based on the multi-bandit theory and random search, respectively. We conduct a thorough empirical evaluation on both simulated and real data. Our proposals can be also viewed as diagnostics for the existence of a reweighted source subsamples that perform better than the random selection of available samples.
Interactive Visualization and Representation Analysis Applied to Glacier Segmentation
Dec 11, 2021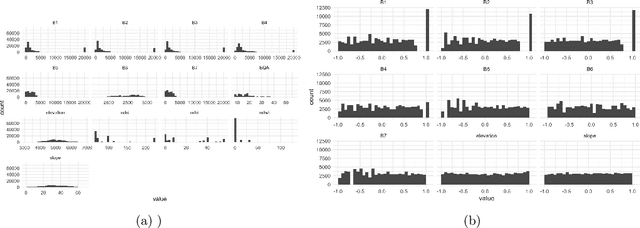


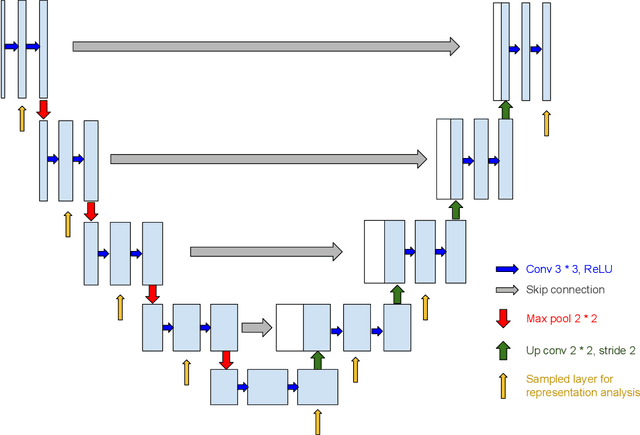
Abstract:Interpretability has attracted increasing attention in earth observation problems. We apply interactive visualization and representation analysis to guide interpretation of glacier segmentation models. We visualize the activations from a U-Net to understand and evaluate the model performance. We build an online interface using the Shiny R package to provide comprehensive error analysis of the predictions. Users can interact with the panels and discover model failure modes. Further, we discuss how visualization can provide sanity checks during data preprocessing and model training.
Interpretability of a Deep Learning Model in the Application of Cardiac MRI Segmentation with an ACDC Challenge Dataset
Mar 15, 2021
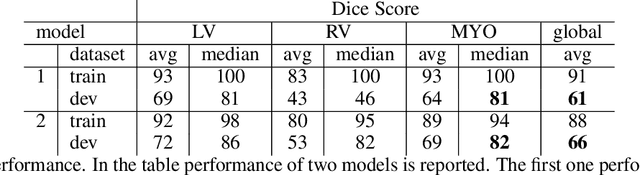

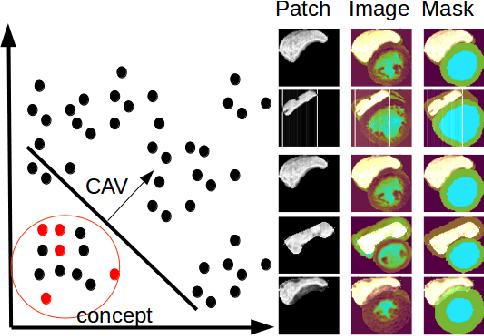
Abstract:Cardiac Magnetic Resonance (CMR) is the most effective tool for the assessment and diagnosis of a heart condition, which malfunction is the world's leading cause of death. Software tools leveraging Artificial Intelligence already enhance radiologists and cardiologists in heart condition assessment but their lack of transparency is a problem. This project investigates if it is possible to discover concepts representative for different cardiac conditions from the deep network trained to segment crdiac structures: Left Ventricle (LV), Right Ventricle (RV) and Myocardium (MYO), using explainability methods that enhances classification system by providing the score-based values of qualitative concepts, along with the key performance metrics. With introduction of a need of explanations in GDPR explainability of AI systems is necessary. This study applies Discovering and Testing with Concept Activation Vectors (D-TCAV), an interpretaibilty method to extract underlying features important for cardiac disease diagnosis from MRI data. The method provides a quantitative notion of concept importance for disease classified. In previous studies, the base method is applied to the classification of cardiac disease and provides clinically meaningful explanations for the predictions of a black-box deep learning classifier. This study applies a method extending TCAV with a Discovering phase (D-TCAV) to cardiac MRI analysis. The advantage of the D-TCAV method over the base method is that it is user-independent. The contribution of this study is a novel application of the explainability method D-TCAV for cardiac MRI anlysis. D-TCAV provides a shorter pre-processing time for clinicians than the base method.
Machine Learning for Glacier Monitoring in the Hindu Kush Himalaya
Dec 09, 2020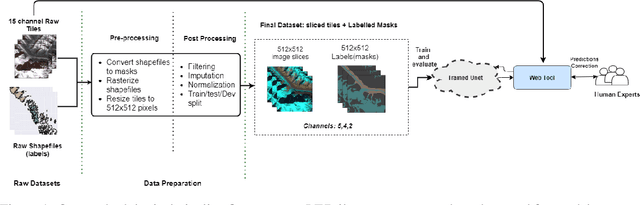
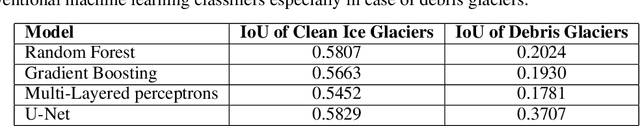
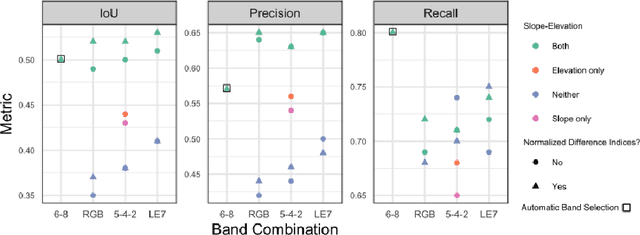
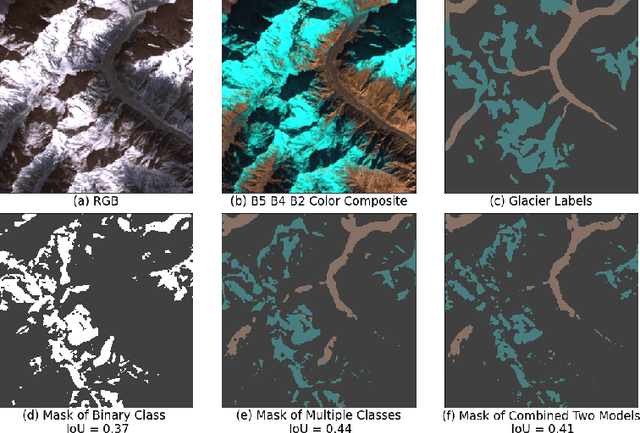
Abstract:Glacier mapping is key to ecological monitoring in the hkh region. Climate change poses a risk to individuals whose livelihoods depend on the health of glacier ecosystems. In this work, we present a machine learning based approach to support ecological monitoring, with a focus on glaciers. Our approach is based on semi-automated mapping from satellite images. We utilize readily available remote sensing data to create a model to identify and outline both clean ice and debris-covered glaciers from satellite imagery. We also release data and develop a web tool that allows experts to visualize and correct model predictions, with the ultimate aim of accelerating the glacier mapping process.
HighRes-net: Recursive Fusion for Multi-Frame Super-Resolution of Satellite Imagery
Feb 15, 2020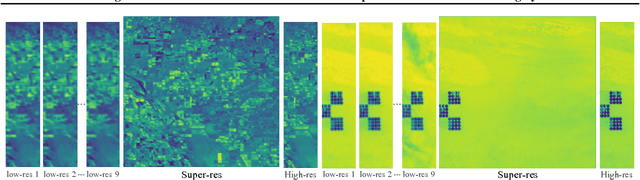
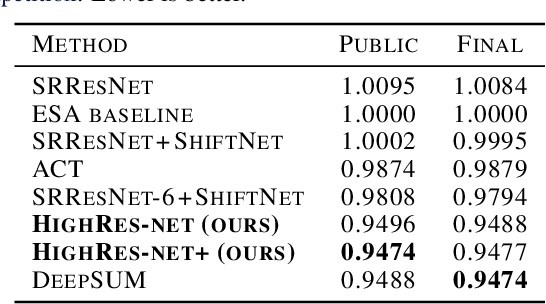
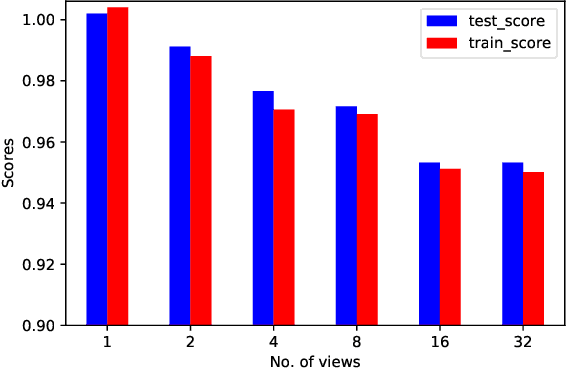
Abstract:Generative deep learning has sparked a new wave of Super-Resolution (SR) algorithms that enhance single images with impressive aesthetic results, albeit with imaginary details. Multi-frame Super-Resolution (MFSR) offers a more grounded approach to the ill-posed problem, by conditioning on multiple low-resolution views. This is important for satellite monitoring of human impact on the planet -- from deforestation, to human rights violations -- that depend on reliable imagery. To this end, we present HighRes-net, the first deep learning approach to MFSR that learns its sub-tasks in an end-to-end fashion: (i) co-registration, (ii) fusion, (iii) up-sampling, and (iv) registration-at-the-loss. Co-registration of low-resolution views is learned implicitly through a reference-frame channel, with no explicit registration mechanism. We learn a global fusion operator that is applied recursively on an arbitrary number of low-resolution pairs. We introduce a registered loss, by learning to align the SR output to a ground-truth through ShiftNet. We show that by learning deep representations of multiple views, we can super-resolve low-resolution signals and enhance Earth Observation data at scale. Our approach recently topped the European Space Agency's MFSR competition on real-world satellite imagery.
Nanoscale Microscopy Images Colourisation Using Neural Networks
Dec 17, 2019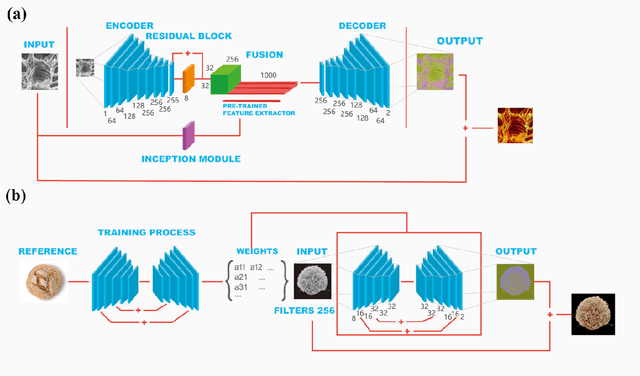
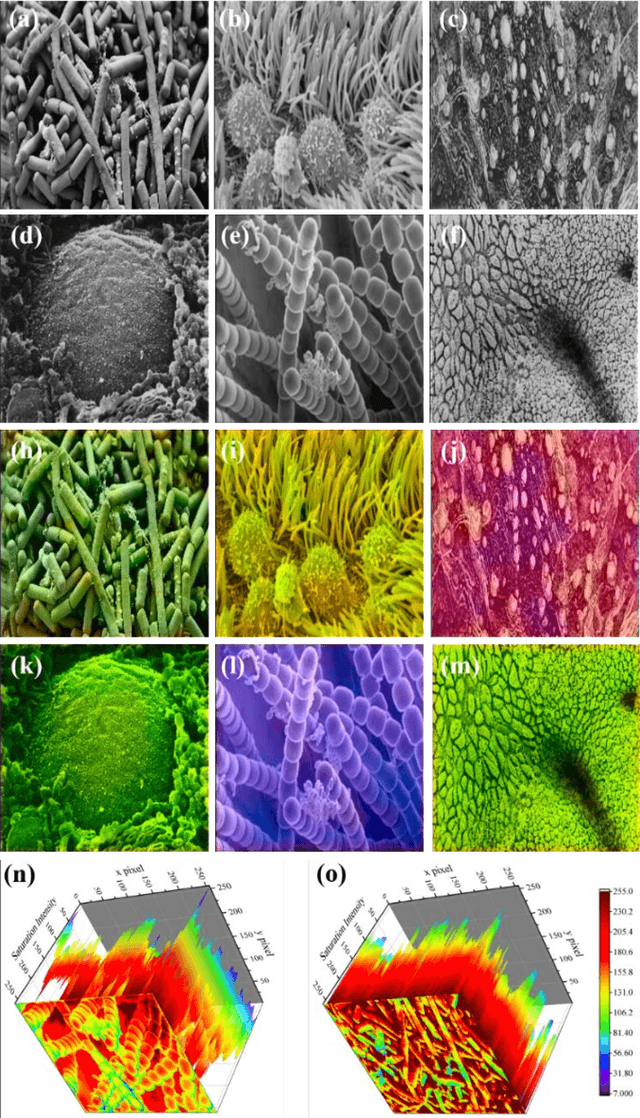

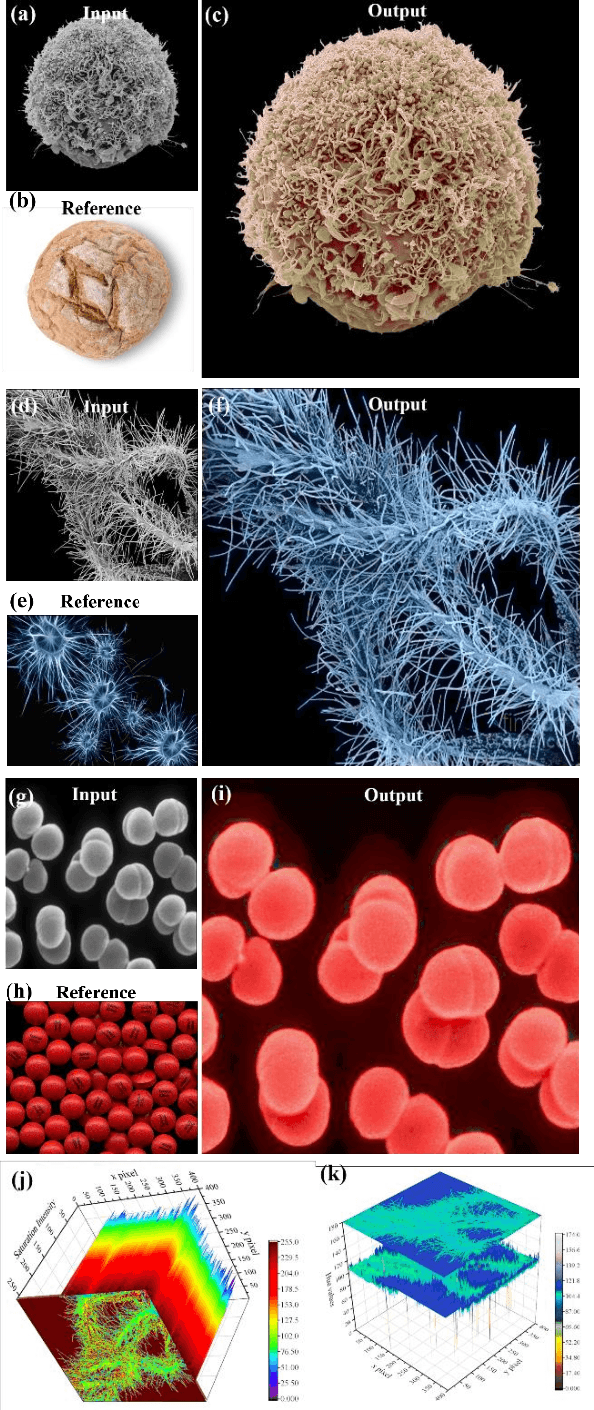
Abstract:Microscopy images are powerful tools and widely used in the majority of research areas, such as biology, chemistry, physics and materials fields by various microscopies (Scanning Electron Microscope (SEM), Atomic Force Microscope (AFM) and the optical microscope, et al.). However, most of the microscopy images are colourless due to the unique imaging mechanism. Though investigating on some popular solutions proposed recently about colourizing microscopy images, we notice the process of those methods are usually tedious, complicated, and time-consuming. In this paper, inspired by the achievement of machine learning algorithms on different science fields, we introduce two artificial neural networks for grey microscopy image colourization: An end-to-end convolutional neural network (CNN) with a pre-trained model for feature extraction and a pixel-to-pixel Neural Style Transfer convolutional neural network (NST-CNN) which can colourize grey microscopy images with semantic information learned from a user-provided colour image at inference time. Our results show that our algorithm not only could able to colour the microscopy images under complex circumstances precisely but also make the colour naturally according to a massive number of nature images training with proper hue and saturation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge