Juanying Xie
Why is the winner the best?
Mar 30, 2023
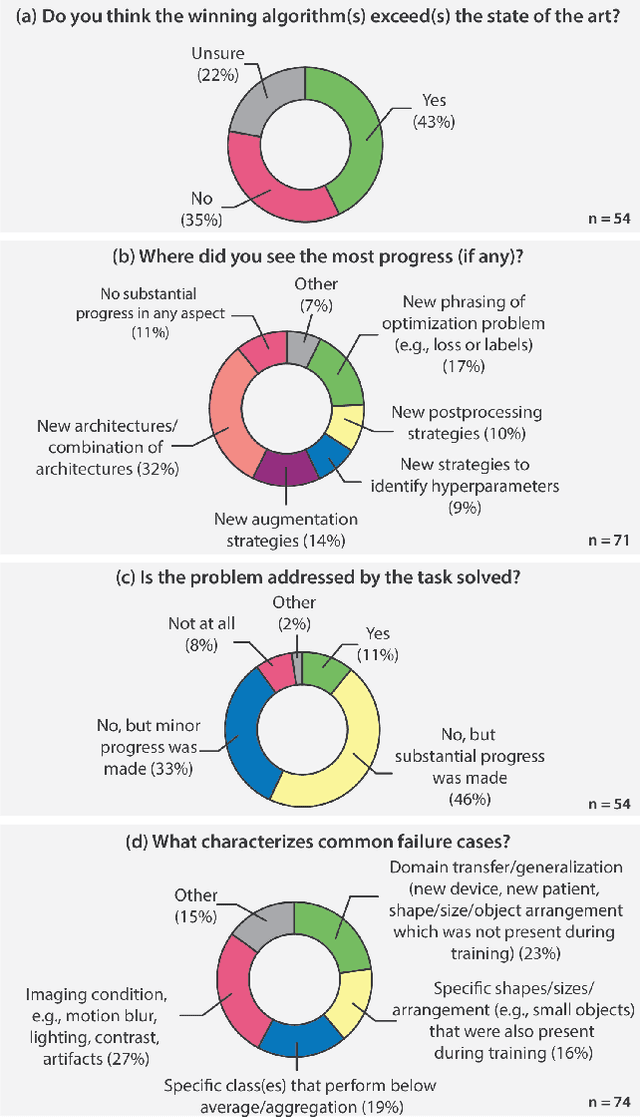
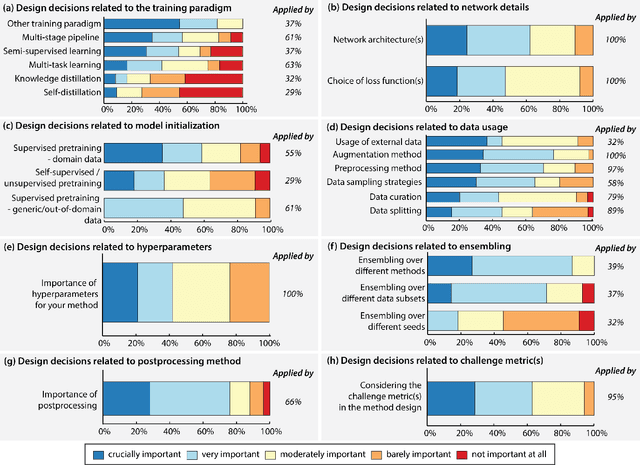
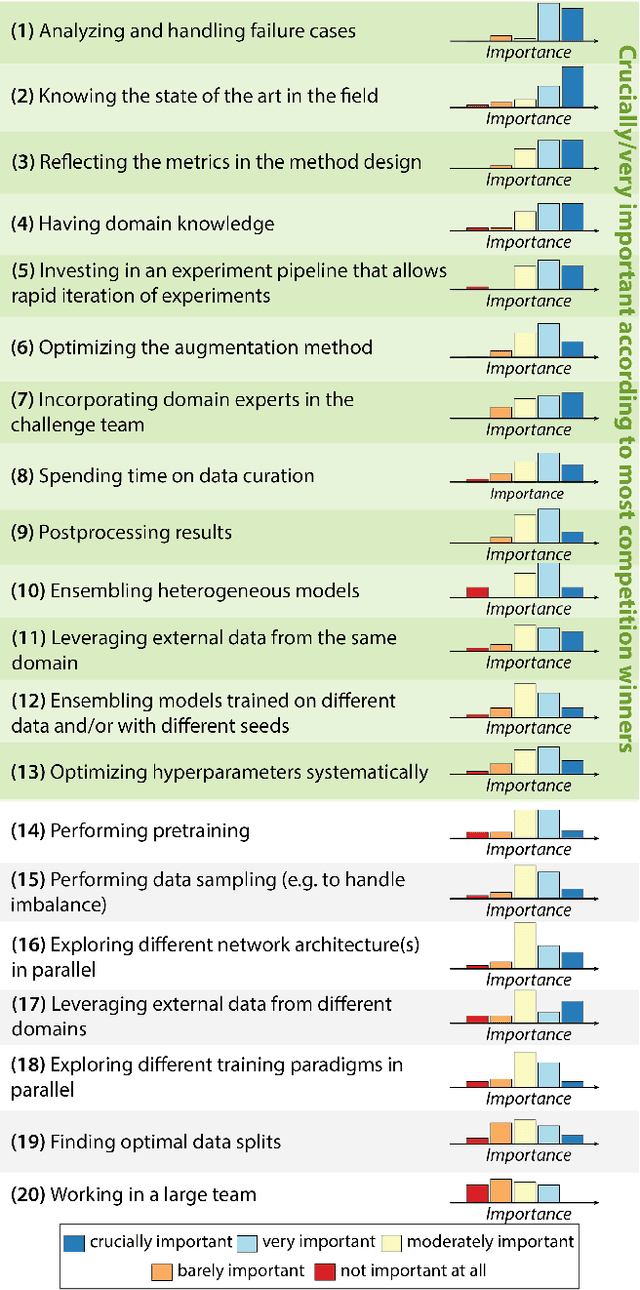
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Fetal Brain Tissue Annotation and Segmentation Challenge Results
Apr 20, 2022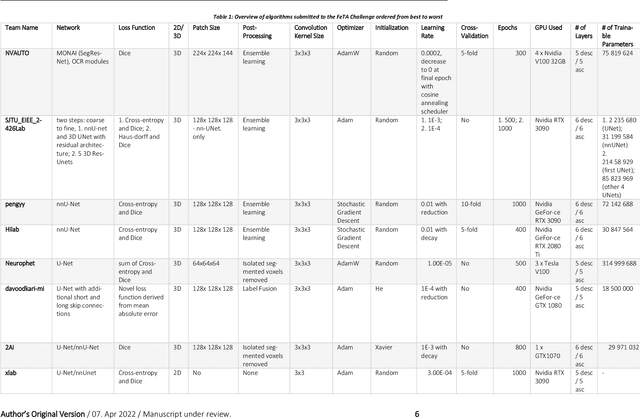
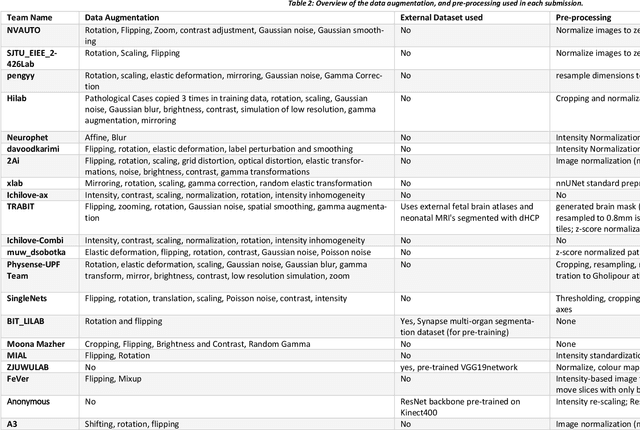
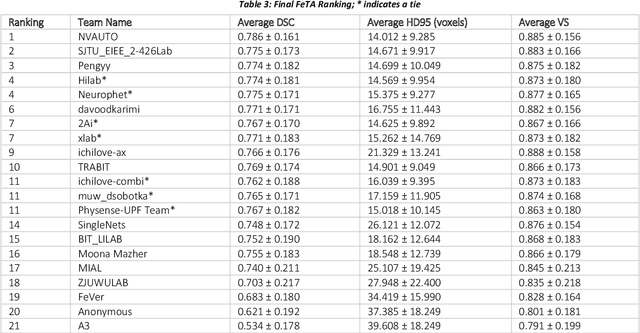
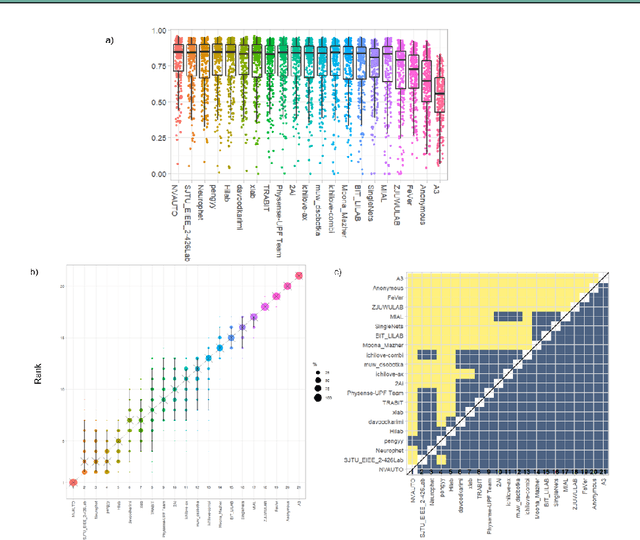
Abstract:In-utero fetal MRI is emerging as an important tool in the diagnosis and analysis of the developing human brain. Automatic segmentation of the developing fetal brain is a vital step in the quantitative analysis of prenatal neurodevelopment both in the research and clinical context. However, manual segmentation of cerebral structures is time-consuming and prone to error and inter-observer variability. Therefore, we organized the Fetal Tissue Annotation (FeTA) Challenge in 2021 in order to encourage the development of automatic segmentation algorithms on an international level. The challenge utilized FeTA Dataset, an open dataset of fetal brain MRI reconstructions segmented into seven different tissues (external cerebrospinal fluid, grey matter, white matter, ventricles, cerebellum, brainstem, deep grey matter). 20 international teams participated in this challenge, submitting a total of 21 algorithms for evaluation. In this paper, we provide a detailed analysis of the results from both a technical and clinical perspective. All participants relied on deep learning methods, mainly U-Nets, with some variability present in the network architecture, optimization, and image pre- and post-processing. The majority of teams used existing medical imaging deep learning frameworks. The main differences between the submissions were the fine tuning done during training, and the specific pre- and post-processing steps performed. The challenge results showed that almost all submissions performed similarly. Four of the top five teams used ensemble learning methods. However, one team's algorithm performed significantly superior to the other submissions, and consisted of an asymmetrical U-Net network architecture. This paper provides a first of its kind benchmark for future automatic multi-tissue segmentation algorithms for the developing human brain in utero.
The Automatic Identification of Butterfly Species
Mar 18, 2018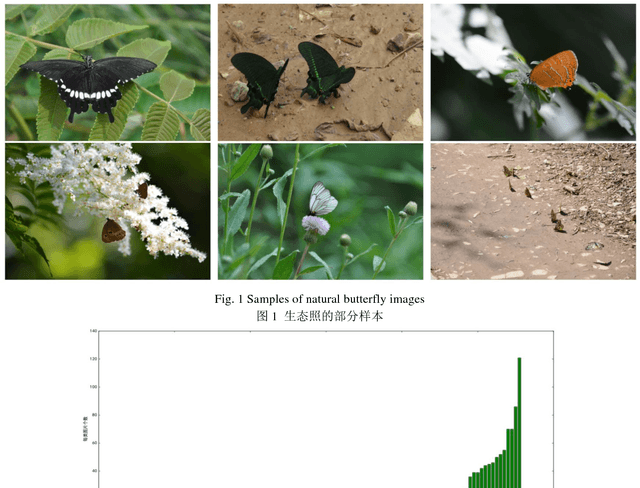
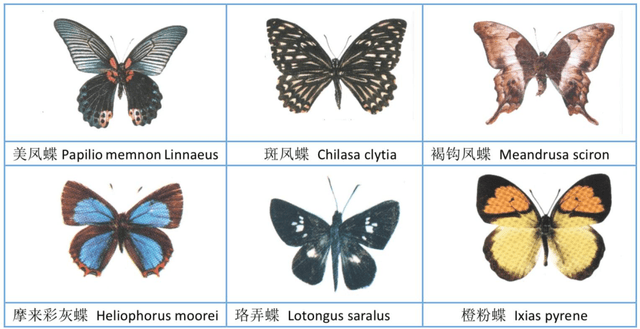
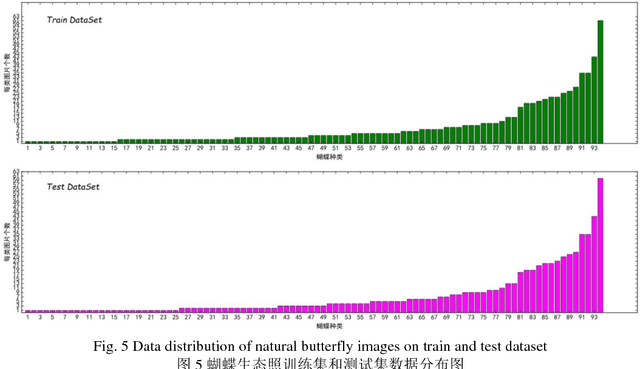
Abstract:The available butterfly data sets comprise a few limited species, and the images in the data sets are always standard patterns without the images of butterflies in their living environment. To overcome the aforementioned limitations in the butterfly data sets, we build a butterfly data set composed of all species of butterflies in China with 4270 standard pattern images of 1176 butterfly species, and 1425 images from living environment of 111 species. We propose to use the deep learning technique Faster-Rcnn to train an automatic butterfly identification system including butterfly position detection and species recognition. We delete those species with only one living environment image from data set, then partition the rest images from living environment into two subsets, one used as test subset, the other as training subset respectively combined with all standard pattern butterfly images or the standard pattern butterfly images with the same species of the images from living environment. In order to construct the training subset for FasterRcnn, nine methods were adopted to amplifying the images in the training subset including the turning of up and down, and left and right, rotation with different angles, adding noises, blurring, and contrast ratio adjusting etc. Three prediction models were trained. The mAP (Mean Average prediction) criterion was used to evaluate the performance of the prediction model. The experimental results demonstrate that our Faster-Rcnn based butterfly automatic identification system performed well, and its worst mAP is up to 60%, and can simultaneously detect the positions of more than one butterflies in one images from living environment and recognize the species of those butterflies as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge