Jing Feng
Uncovering What, Why and How: A Comprehensive Benchmark for Causation Understanding of Video Anomaly
Apr 30, 2024



Abstract:Video anomaly understanding (VAU) aims to automatically comprehend unusual occurrences in videos, thereby enabling various applications such as traffic surveillance and industrial manufacturing. While existing VAU benchmarks primarily concentrate on anomaly detection and localization, our focus is on more practicality, prompting us to raise the following crucial questions: "what anomaly occurred?", "why did it happen?", and "how severe is this abnormal event?". In pursuit of these answers, we present a comprehensive benchmark for Causation Understanding of Video Anomaly (CUVA). Specifically, each instance of the proposed benchmark involves three sets of human annotations to indicate the "what", "why" and "how" of an anomaly, including 1) anomaly type, start and end times, and event descriptions, 2) natural language explanations for the cause of an anomaly, and 3) free text reflecting the effect of the abnormality. In addition, we also introduce MMEval, a novel evaluation metric designed to better align with human preferences for CUVA, facilitating the measurement of existing LLMs in comprehending the underlying cause and corresponding effect of video anomalies. Finally, we propose a novel prompt-based method that can serve as a baseline approach for the challenging CUVA. We conduct extensive experiments to show the superiority of our evaluation metric and the prompt-based approach. Our code and dataset are available at https://github.com/fesvhtr/CUVA.
Deeply Supervised Layer Selective Attention Network: Towards Label-Efficient Learning for Medical Image Classification
Sep 28, 2022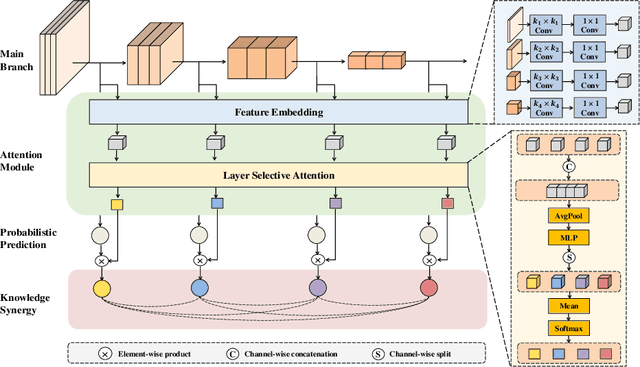
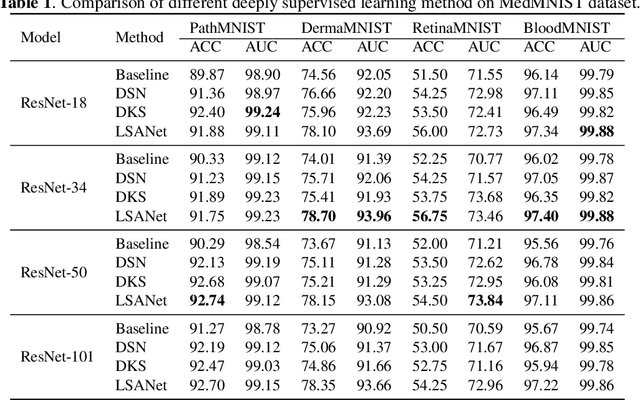
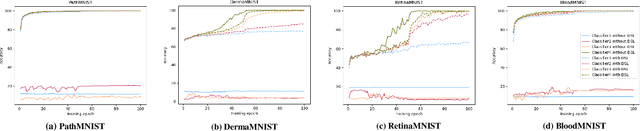
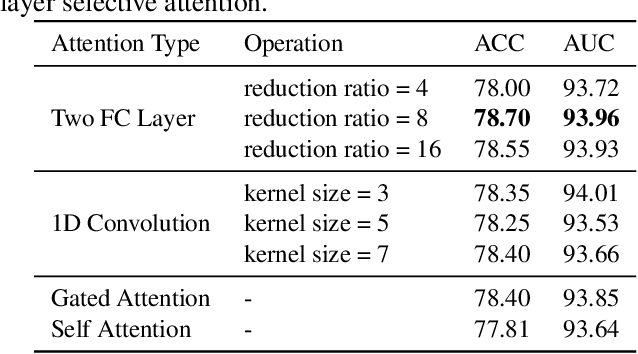
Abstract:Labeling medical images depends on professional knowledge, making it difficult to acquire large amount of annotated medical images with high quality in a short time. Thus, making good use of limited labeled samples in a small dataset to build a high-performance model is the key to medical image classification problem. In this paper, we propose a deeply supervised Layer Selective Attention Network (LSANet), which comprehensively uses label information in feature-level and prediction-level supervision. For feature-level supervision, in order to better fuse the low-level features and high-level features, we propose a novel visual attention module, Layer Selective Attention (LSA), to focus on the feature selection of different layers. LSA introduces a weight allocation scheme which can dynamically adjust the weighting factor of each auxiliary branch during the whole training process to further enhance deeply supervised learning and ensure its generalization. For prediction-level supervision, we adopt the knowledge synergy strategy to promote hierarchical information interactions among all supervision branches via pairwise knowledge matching. Using the public dataset, MedMNIST, which is a large-scale benchmark for biomedical image classification covering diverse medical specialties, we evaluate LSANet on multiple mainstream CNN architectures and various visual attention modules. The experimental results show the substantial improvements of our proposed method over its corresponding counterparts, demonstrating that LSANet can provide a promising solution for label-efficient learning in the field of medical image classification.
DrugOOD: Out-of-Distribution (OOD) Dataset Curator and Benchmark for AI-aided Drug Discovery -- A Focus on Affinity Prediction Problems with Noise Annotations
Jan 24, 2022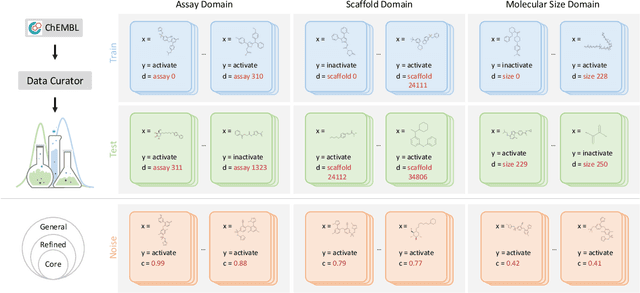
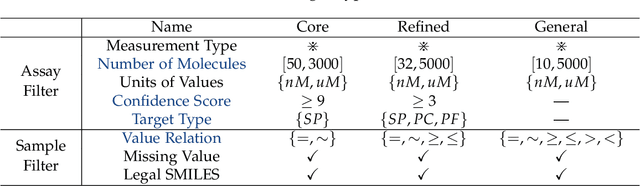
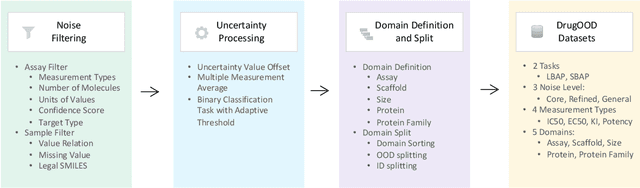
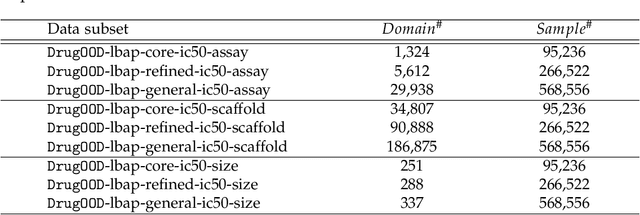
Abstract:AI-aided drug discovery (AIDD) is gaining increasing popularity due to its promise of making the search for new pharmaceuticals quicker, cheaper and more efficient. In spite of its extensive use in many fields, such as ADMET prediction, virtual screening, protein folding and generative chemistry, little has been explored in terms of the out-of-distribution (OOD) learning problem with \emph{noise}, which is inevitable in real world AIDD applications. In this work, we present DrugOOD, a systematic OOD dataset curator and benchmark for AI-aided drug discovery, which comes with an open-source Python package that fully automates the data curation and OOD benchmarking processes. We focus on one of the most crucial problems in AIDD: drug target binding affinity prediction, which involves both macromolecule (protein target) and small-molecule (drug compound). In contrast to only providing fixed datasets, DrugOOD offers automated dataset curator with user-friendly customization scripts, rich domain annotations aligned with biochemistry knowledge, realistic noise annotations and rigorous benchmarking of state-of-the-art OOD algorithms. Since the molecular data is often modeled as irregular graphs using graph neural network (GNN) backbones, DrugOOD also serves as a valuable testbed for \emph{graph OOD learning} problems. Extensive empirical studies have shown a significant performance gap between in-distribution and out-of-distribution experiments, which highlights the need to develop better schemes that can allow for OOD generalization under noise for AIDD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge