Hongyu He
AI-generated data contamination erodes pathological variability and diagnostic reliability
Jan 21, 2026Abstract:Generative artificial intelligence (AI) is rapidly populating medical records with synthetic content, creating a feedback loop where future models are increasingly at risk of training on uncurated AI-generated data. However, the clinical consequences of this AI-generated data contamination remain unexplored. Here, we show that in the absence of mandatory human verification, this self-referential cycle drives a rapid erosion of pathological variability and diagnostic reliability. By analysing more than 800,000 synthetic data points across clinical text generation, vision-language reporting, and medical image synthesis, we find that models progressively converge toward generic phenotypes regardless of the model architecture. Specifically, rare but critical findings, including pneumothorax and effusions, vanish from the synthetic content generated by AI models, while demographic representations skew heavily toward middle-aged male phenotypes. Crucially, this degradation is masked by false diagnostic confidence; models continue to issue reassuring reports while failing to detect life-threatening pathology, with false reassurance rates tripling to 40%. Blinded physician evaluation confirms that this decoupling of confidence and accuracy renders AI-generated documentation clinically useless after just two generations. We systematically evaluate three mitigation strategies, finding that while synthetic volume scaling fails to prevent collapse, mixing real data with quality-aware filtering effectively preserves diversity. Ultimately, our results suggest that without policy-mandated human oversight, the deployment of generative AI threatens to degrade the very healthcare data ecosystems it relies upon.
LLMvsSmall Model? Large Language Model Based Text Augmentation Enhanced Personality Detection Model
Mar 12, 2024Abstract:Personality detection aims to detect one's personality traits underlying in social media posts. One challenge of this task is the scarcity of ground-truth personality traits which are collected from self-report questionnaires. Most existing methods learn post features directly by fine-tuning the pre-trained language models under the supervision of limited personality labels. This leads to inferior quality of post features and consequently affects the performance. In addition, they treat personality traits as one-hot classification labels, overlooking the semantic information within them. In this paper, we propose a large language model (LLM) based text augmentation enhanced personality detection model, which distills the LLM's knowledge to enhance the small model for personality detection, even when the LLM fails in this task. Specifically, we enable LLM to generate post analyses (augmentations) from the aspects of semantic, sentiment, and linguistic, which are critical for personality detection. By using contrastive learning to pull them together in the embedding space, the post encoder can better capture the psycho-linguistic information within the post representations, thus improving personality detection. Furthermore, we utilize the LLM to enrich the information of personality labels for enhancing the detection performance. Experimental results on the benchmark datasets demonstrate that our model outperforms the state-of-the-art methods on personality detection.
SuperMask: Generating High-resolution object masks from multi-view, unaligned low-resolution MRIs
Mar 13, 2023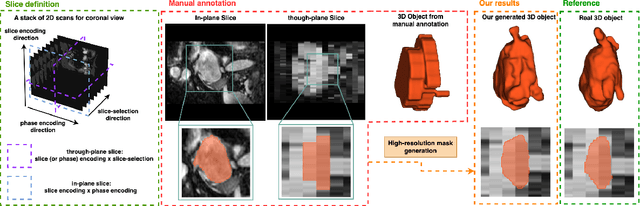

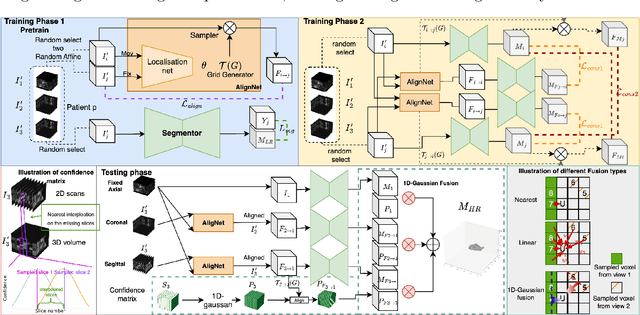
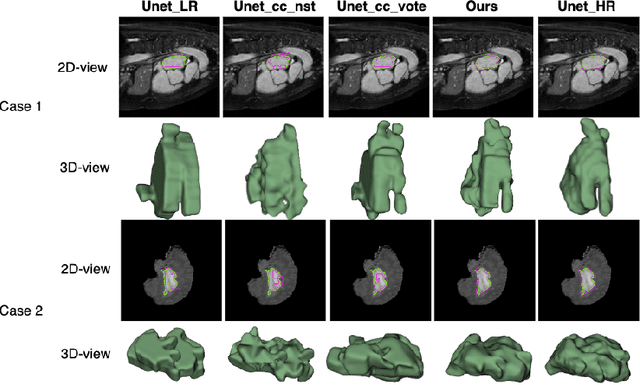
Abstract:Three-dimensional segmentation in magnetic resonance images (MRI), which reflects the true shape of the objects, is challenging since high-resolution isotropic MRIs are rare and typical MRIs are anisotropic, with the out-of-plane dimension having a much lower resolution. A potential remedy to this issue lies in the fact that often multiple sequences are acquired on different planes. However, in practice, these sequences are not orthogonal to each other, limiting the applicability of many previous solutions to reconstruct higher-resolution images from multiple lower-resolution ones. We propose a weakly-supervised deep learning-based solution to generating high-resolution masks from multiple low-resolution images. Our method combines segmentation and unsupervised registration networks by introducing two new regularizations to make registration and segmentation reinforce each other. Finally, we introduce a multi-view fusion method to generate high-resolution target object masks. The experimental results on two datasets show the superiority of our methods. Importantly, the advantage of not using high-resolution images in the training process makes our method applicable to a wide variety of MRI segmentation tasks.
FaceGuard: Proactive Deepfake Detection
Sep 13, 2021
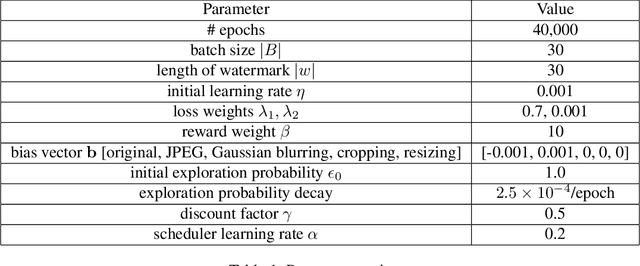


Abstract:Existing deepfake-detection methods focus on passive detection, i.e., they detect fake face images via exploiting the artifacts produced during deepfake manipulation. A key limitation of passive detection is that it cannot detect fake faces that are generated by new deepfake generation methods. In this work, we propose FaceGuard, a proactive deepfake-detection framework. FaceGuard embeds a watermark into a real face image before it is published on social media. Given a face image that claims to be an individual (e.g., Nicolas Cage), FaceGuard extracts a watermark from it and predicts the face image to be fake if the extracted watermark does not match well with the individual's ground truth one. A key component of FaceGuard is a new deep-learning-based watermarking method, which is 1) robust to normal image post-processing such as JPEG compression, Gaussian blurring, cropping, and resizing, but 2) fragile to deepfake manipulation. Our evaluation on multiple datasets shows that FaceGuard can detect deepfakes accurately and outperforms existing methods.
Tab2Know: Building a Knowledge Base from Tables in Scientific Papers
Jul 28, 2021



Abstract:Tables in scientific papers contain a wealth of valuable knowledge for the scientific enterprise. To help the many of us who frequently consult this type of knowledge, we present Tab2Know, a new end-to-end system to build a Knowledge Base (KB) from tables in scientific papers. Tab2Know addresses the challenge of automatically interpreting the tables in papers and of disambiguating the entities that they contain. To solve these problems, we propose a pipeline that employs both statistical-based classifiers and logic-based reasoning. First, our pipeline applies weakly supervised classifiers to recognize the type of tables and columns, with the help of a data labeling system and an ontology specifically designed for our purpose. Then, logic-based reasoning is used to link equivalent entities (via sameAs links) in different tables. An empirical evaluation of our approach using a corpus of papers in the Computer Science domain has returned satisfactory performance. This suggests that ours is a promising step to create a large-scale KB of scientific knowledge.
* 17 pages, 4 figures, conference: The Semantic Web -- ISWC 2020
OpenHI2 -- Open source histopathological image platform
Jan 15, 2020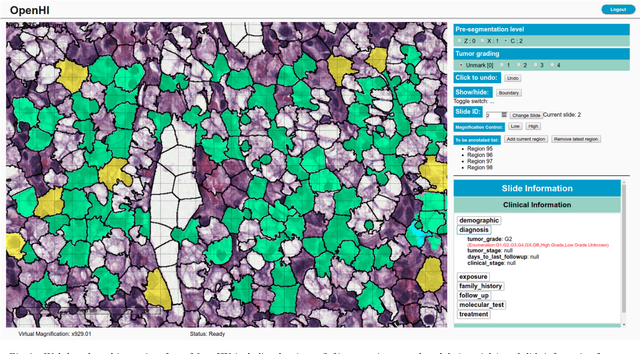
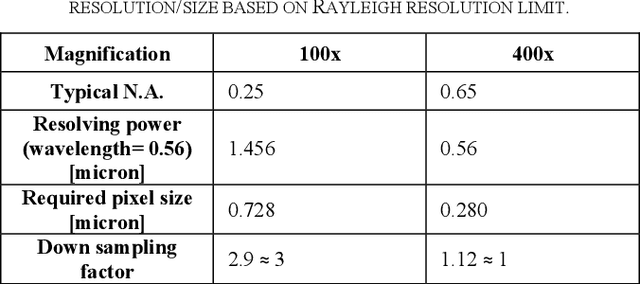
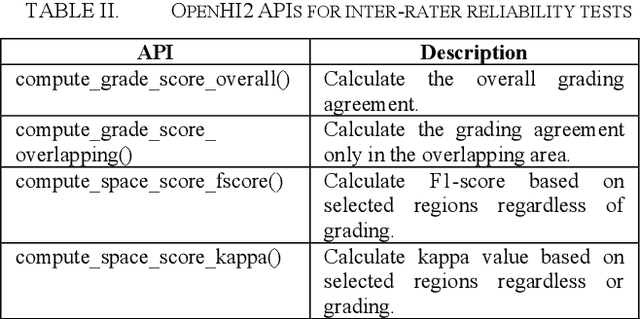
Abstract:Transition from conventional to digital pathology requires a new category of biomedical informatic infrastructure which could facilitate delicate pathological routine. Pathological diagnoses are sensitive to many external factors and is known to be subjective. Only systems that can meet strict requirements in pathology would be able to run along pathological routines and eventually digitized the study area, and the developed platform should comply with existing pathological routines and international standards. Currently, there are a number of available software tools which can perform histopathological tasks including virtual slide viewing, annotating, and basic image analysis, however, none of them can serve as a digital platform for pathology. Here we describe OpenHI2, an enhanced version Open Histopathological Image platform which is capable of supporting all basic pathological tasks and file formats; ready to be deployed in medical institutions on a standard server environment or cloud computing infrastructure. In this paper, we also describe the development decisions for the platform and propose solutions to overcome technical challenges so that OpenHI2 could be used as a platform for histopathological images. Further addition can be made to the platform since each component is modularized and fully documented. OpenHI2 is free, open-source, and available at https://gitlab.com/BioAI/OpenHI.
Stochastic Channel-Based Federated Learning for Medical Data Privacy Preserving
Nov 15, 2019

Abstract:Artificial neural network has achieved unprecedented success in the medical domain. This success depends on the availability of massive and representative datasets. However, data collection is often prevented by privacy concerns and people want to take control over their sensitive information during both training and using processes. To address this problem, we propose a privacy-preserving method for the distributed system, Stochastic Channel-Based Federated Learning (SCBF), which enables the participants to train a high-performance model cooperatively without sharing their inputs. Specifically, we design, implement and evaluate a channel-based update algorithm for the central server in a distributed system, which selects the channels with regard to the most active features in a training loop and uploads them as learned information from local datasets. A pruning process is applied to the algorithm based on the validation set, which serves as a model accelerator. In the experiment, our model presents better performances and higher saturating speed than the Federated Averaging method which reveals all the parameters of local models to the server when updating. We also demonstrate that the saturating rate of performance could be promoted by introducing a pruning process. And further improvement could be achieved by tuning the pruning rate. Our experiment shows that 57% of the time is saved by the pruning process with only a reduction of 0.0047 in AUCROC performance and a reduction of 0.0068 in AUCPR.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge