Chunbao Wang
Meta Mask Correction for Nuclei Segmentation in Histopathological Image
Nov 24, 2021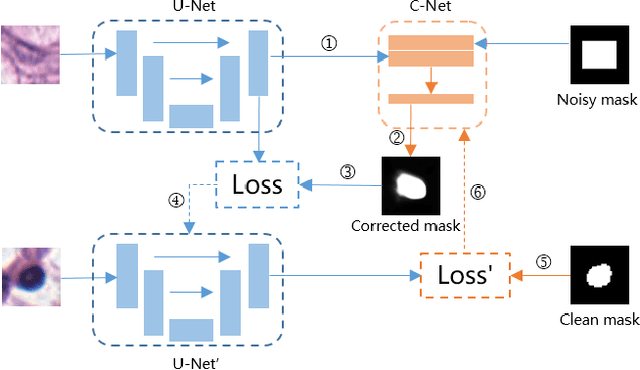
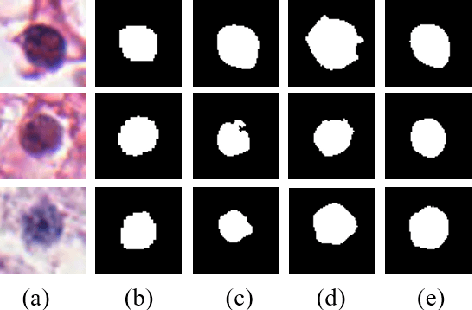
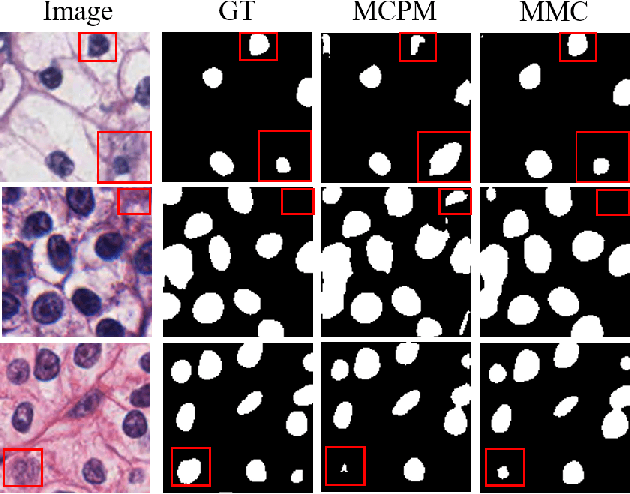
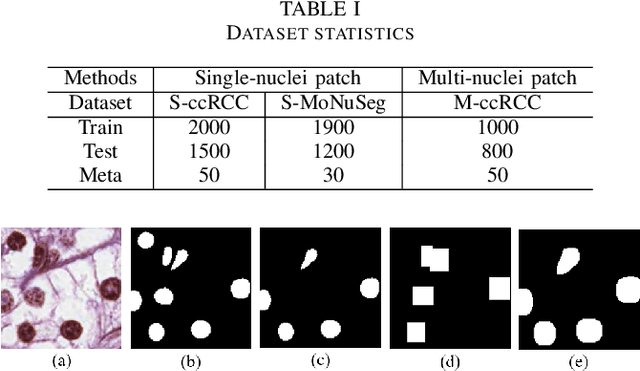
Abstract:Nuclei segmentation is a fundamental task in digital pathology analysis and can be automated by deep learning-based methods. However, the development of such an automated method requires a large amount of data with precisely annotated masks which is hard to obtain. Training with weakly labeled data is a popular solution for reducing the workload of annotation. In this paper, we propose a novel meta-learning-based nuclei segmentation method which follows the label correction paradigm to leverage data with noisy masks. Specifically, we design a fully conventional meta-model that can correct noisy masks using a small amount of clean meta-data. Then the corrected masks can be used to supervise the training of the segmentation model. Meanwhile, a bi-level optimization method is adopted to alternately update the parameters of the main segmentation model and the meta-model in an end-to-end way. Extensive experimental results on two nuclear segmentation datasets show that our method achieves the state-of-the-art result. It even achieves comparable performance with the model training on supervised data in some noisy settings.
PIMIP: An Open Source Platform for Pathology Information Management and Integration
Nov 09, 2021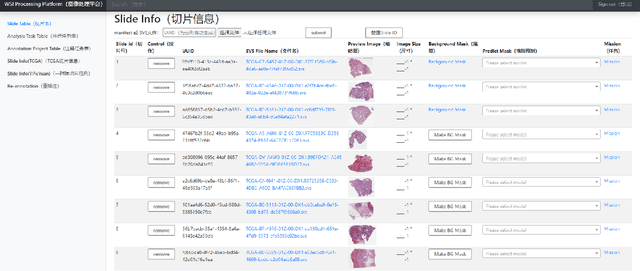
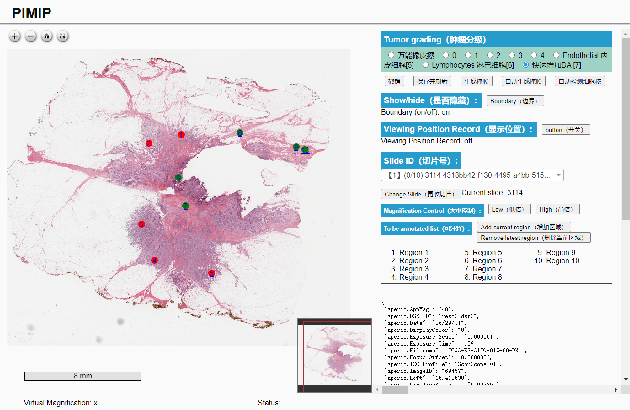
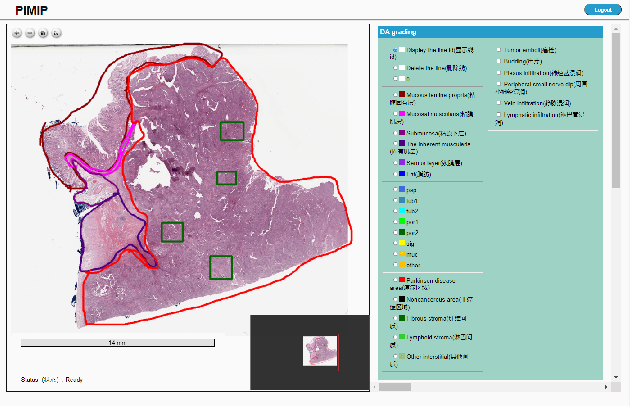
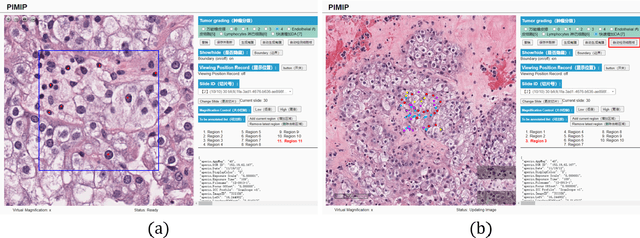
Abstract:Digital pathology plays a crucial role in the development of artificial intelligence in the medical field. The digital pathology platform can make the pathological resources digital and networked, and realize the permanent storage of visual data and the synchronous browsing processing without the limitation of time and space. It has been widely used in various fields of pathology. However, there is still a lack of an open and universal digital pathology platform to assist doctors in the management and analysis of digital pathological sections, as well as the management and structured description of relevant patient information. Most platforms cannot integrate image viewing, annotation and analysis, and text information management. To solve the above problems, we propose a comprehensive and extensible platform PIMIP. Our PIMIP has developed the image annotation functions based on the visualization of digital pathological sections. Our annotation functions support multi-user collaborative annotation and multi-device annotation, and realize the automation of some annotation tasks. In the annotation task, we invited a professional pathologist for guidance. We introduce a machine learning module for image analysis. The data we collected included public data from local hospitals and clinical examples. Our platform is more clinical and suitable for clinical use. In addition to image data, we also structured the management and display of text information. So our platform is comprehensive. The platform framework is built in a modular way to support users to add machine learning modules independently, which makes our platform extensible.
BioIE: Biomedical Information Extraction with Multi-head Attention Enhanced Graph Convolutional Network
Oct 26, 2021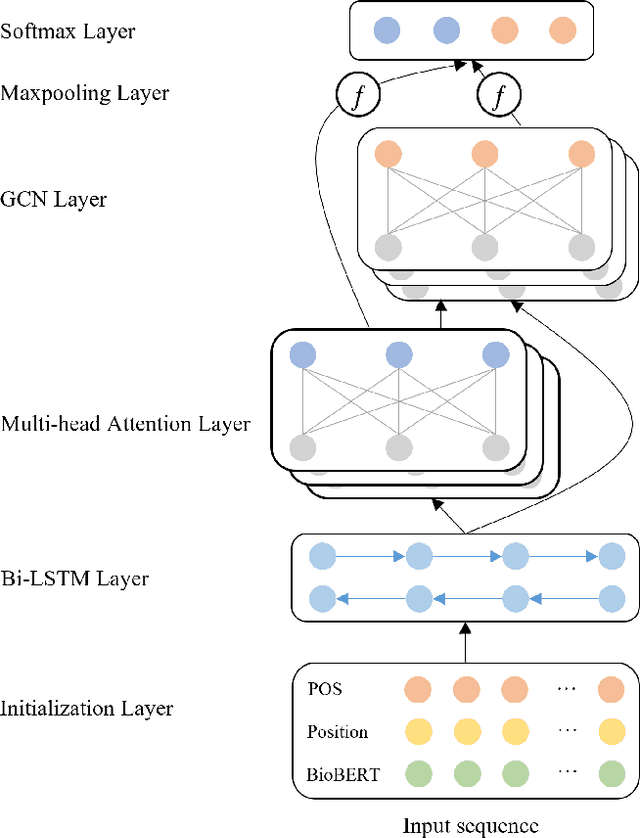
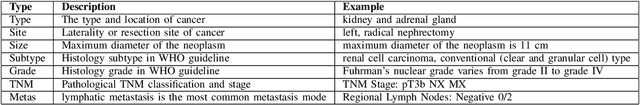
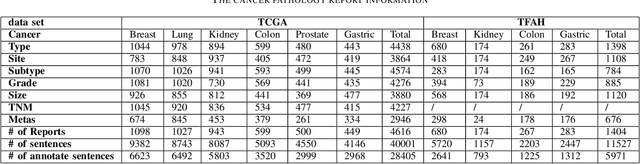
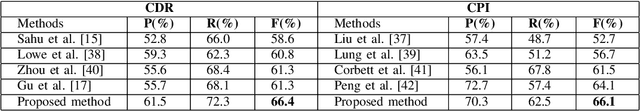
Abstract:Constructing large-scaled medical knowledge graphs can significantly boost healthcare applications for medical surveillance, bring much attention from recent research. An essential step in constructing large-scale MKG is extracting information from medical reports. Recently, information extraction techniques have been proposed and show promising performance in biomedical information extraction. However, these methods only consider limited types of entity and relation due to the noisy biomedical text data with complex entity correlations. Thus, they fail to provide enough information for constructing MKGs and restrict the downstream applications. To address this issue, we propose Biomedical Information Extraction, a hybrid neural network to extract relations from biomedical text and unstructured medical reports. Our model utilizes a multi-head attention enhanced graph convolutional network to capture the complex relations and context information while resisting the noise from the data. We evaluate our model on two major biomedical relationship extraction tasks, chemical-disease relation and chemical-protein interaction, and a cross-hospital pan-cancer pathology report corpus. The results show that our method achieves superior performance than baselines. Furthermore, we evaluate the applicability of our method under a transfer learning setting and show that BioIE achieves promising performance in processing medical text from different formats and writing styles.
A Personalized Diagnostic Generation Framework Based on Multi-source Heterogeneous Data
Oct 26, 2021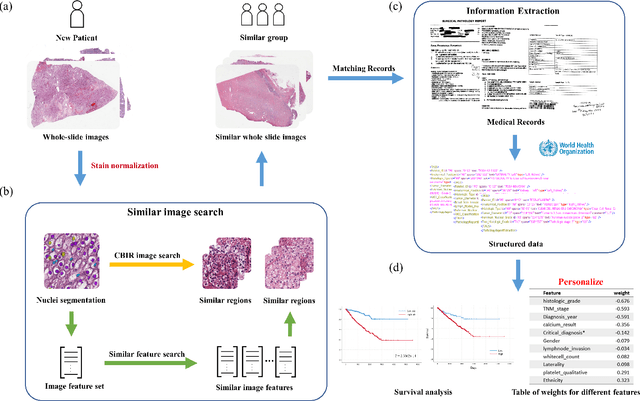
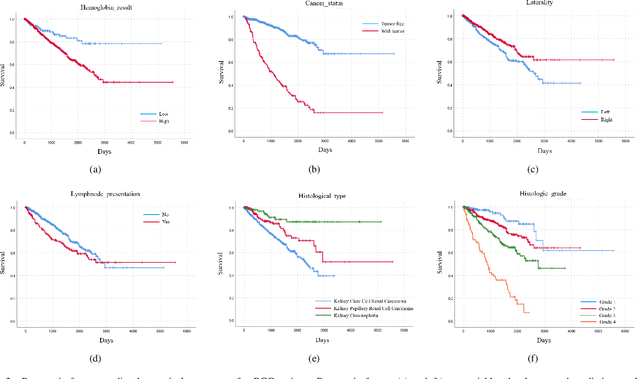
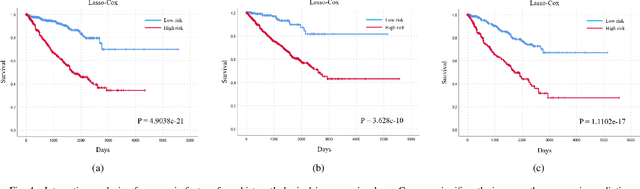
Abstract:Personalized diagnoses have not been possible due to sear amount of data pathologists have to bear during the day-to-day routine. This lead to the current generalized standards that are being continuously updated as new findings are reported. It is noticeable that these effective standards are developed based on a multi-source heterogeneous data, including whole-slide images and pathology and clinical reports. In this study, we propose a framework that combines pathological images and medical reports to generate a personalized diagnosis result for individual patient. We use nuclei-level image feature similarity and content-based deep learning method to search for a personalized group of population with similar pathological characteristics, extract structured prognostic information from descriptive pathology reports of the similar patient population, and assign importance of different prognostic factors to generate a personalized pathological diagnosis result. We use multi-source heterogeneous data from TCGA (The Cancer Genome Atlas) database. The result demonstrate that our framework matches the performance of pathologists in the diagnosis of renal cell carcinoma. This framework is designed to be generic, thus could be applied for other types of cancer. The weights could provide insights to the known prognostic factors and further guide more precise clinical treatment protocols.
A Precision Diagnostic Framework of Renal Cell Carcinoma on Whole-Slide Images using Deep Learning
Oct 26, 2021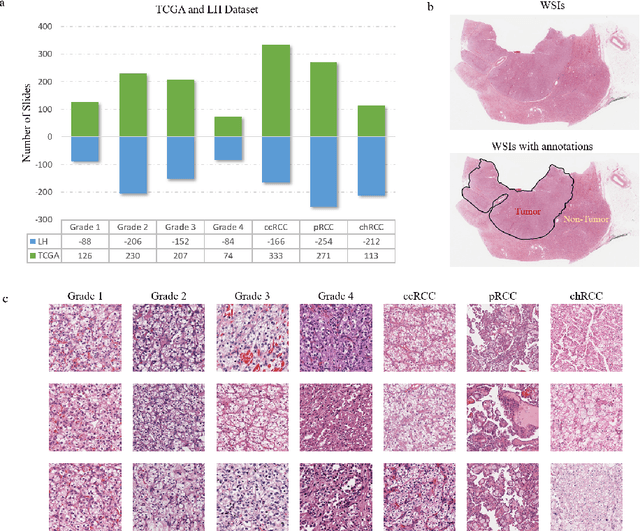
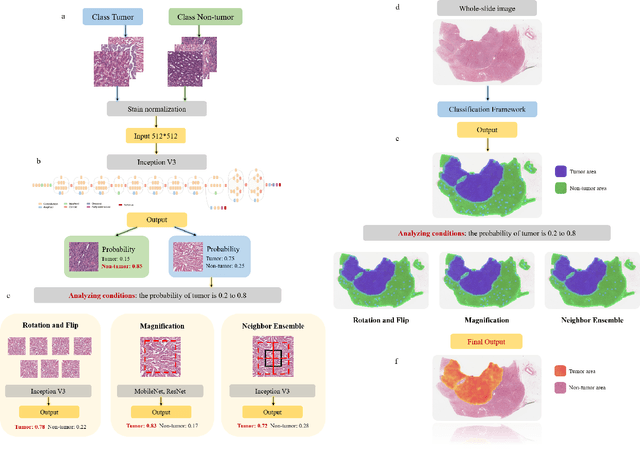
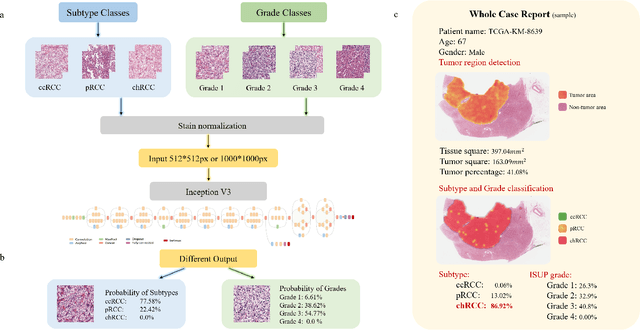
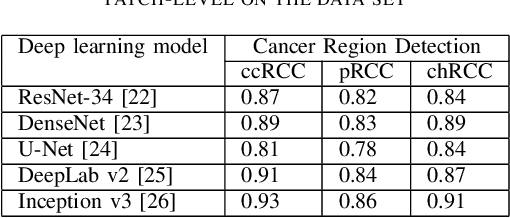
Abstract:Diagnostic pathology, which is the basis and gold standard of cancer diagnosis, provides essential information on the prognosis of the disease and vital evidence for clinical treatment. Tumor region detection, subtype and grade classification are the fundamental diagnostic indicators for renal cell carcinoma (RCC) in whole-slide images (WSIs). However, pathological diagnosis is subjective, differences in observation and diagnosis between pathologists is common in hospitals with inadequate diagnostic capacity. The main challenge for developing deep learning based RCC diagnostic system is the lack of large-scale datasets with precise annotations. In this work, we proposed a deep learning-based framework for analyzing histopathological images of patients with renal cell carcinoma, which has the potential to achieve pathologist-level accuracy in diagnosis. A deep convolutional neural network (InceptionV3) was trained on the high-quality annotated dataset of The Cancer Genome Atlas (TCGA) whole-slide histopathological image for accurate tumor area detection, classification of RCC subtypes, and ISUP grades classification of clear cell carcinoma subtypes. These results suggest that our framework can help pathologists in the detection of cancer region and classification of subtypes and grades, which could be applied to any cancer type, providing auxiliary diagnosis and promoting clinical consensus.
Instance-based Vision Transformer for Subtyping of Papillary Renal Cell Carcinoma in Histopathological Image
Jun 23, 2021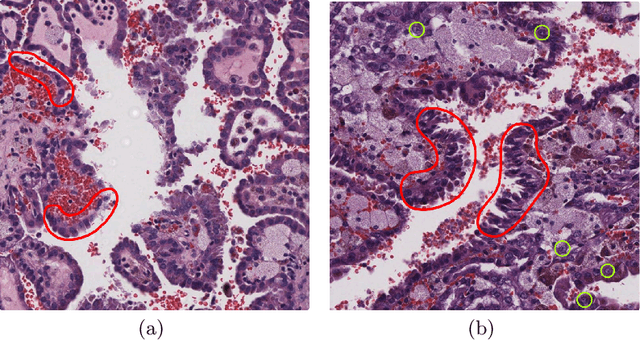
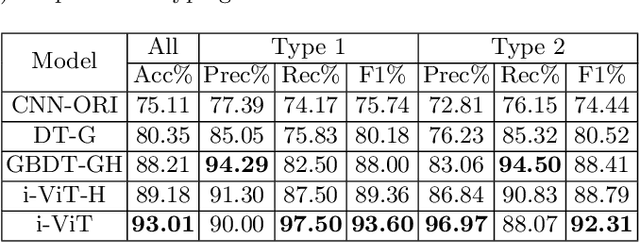
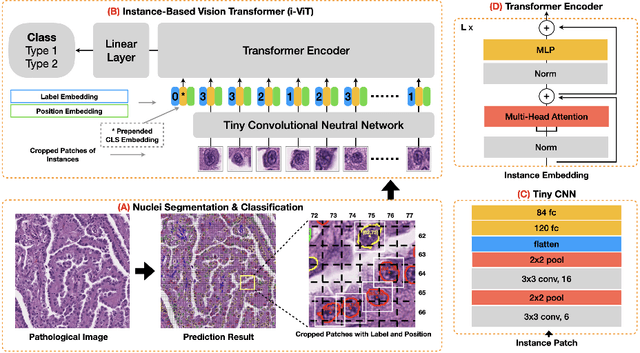
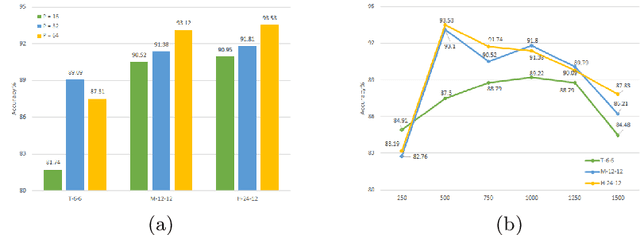
Abstract:Histological subtype of papillary (p) renal cell carcinoma (RCC), type 1 vs. type 2, is an essential prognostic factor. The two subtypes of pRCC have a similar pattern, i.e., the papillary architecture, yet some subtle differences, including cellular and cell-layer level patterns. However, the cellular and cell-layer level patterns almost cannot be captured by existing CNN-based models in large-size histopathological images, which brings obstacles to directly applying these models to such a fine-grained classification task. This paper proposes a novel instance-based Vision Transformer (i-ViT) to learn robust representations of histopathological images for the pRCC subtyping task by extracting finer features from instance patches (by cropping around segmented nuclei and assigning predicted grades). The proposed i-ViT takes top-K instances as input and aggregates them for capturing both the cellular and cell-layer level patterns by a position-embedding layer, a grade-embedding layer, and a multi-head multi-layer self-attention module. To evaluate the performance of the proposed framework, experienced pathologists are invited to selected 1162 regions of interest from 171 whole slide images of type 1 and type 2 pRCC. Experimental results show that the proposed method achieves better performance than existing CNN-based models with a significant margin.
Nuclei Grading of Clear Cell Renal Cell Carcinoma in Histopathological Image by Composite High-Resolution Network
Jun 20, 2021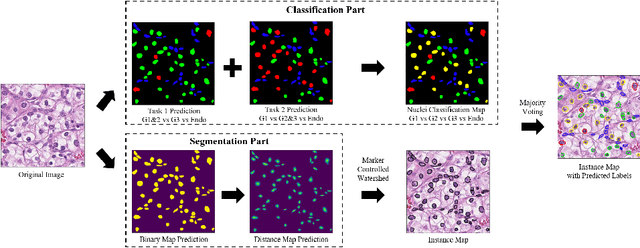
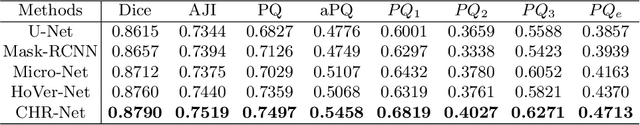
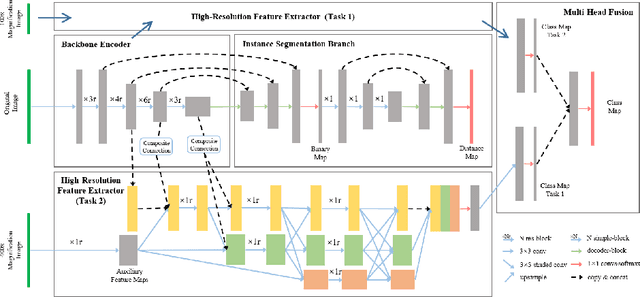
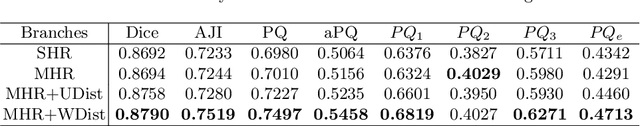
Abstract:The grade of clear cell renal cell carcinoma (ccRCC) is a critical prognostic factor, making ccRCC nuclei grading a crucial task in RCC pathology analysis. Computer-aided nuclei grading aims to improve pathologists' work efficiency while reducing their misdiagnosis rate by automatically identifying the grades of tumor nuclei within histopathological images. Such a task requires precisely segment and accurately classify the nuclei. However, most of the existing nuclei segmentation and classification methods can not handle the inter-class similarity property of nuclei grading, thus can not be directly applied to the ccRCC grading task. In this paper, we propose a Composite High-Resolution Network for ccRCC nuclei grading. Specifically, we propose a segmentation network called W-Net that can separate the clustered nuclei. Then, we recast the fine-grained classification of nuclei to two cross-category classification tasks, based on two high-resolution feature extractors (HRFEs) which are proposed for learning these two tasks. The two HRFEs share the same backbone encoder with W-Net by a composite connection so that meaningful features for the segmentation task can be inherited for the classification task. Last, a head-fusion block is applied to generate the predicted label of each nucleus. Furthermore, we introduce a dataset for ccRCC nuclei grading, containing 1000 image patches with 70945 annotated nuclei. We demonstrate that our proposed method achieves state-of-the-art performance compared to existing methods on this large ccRCC grading dataset.
OpenHI2 -- Open source histopathological image platform
Jan 15, 2020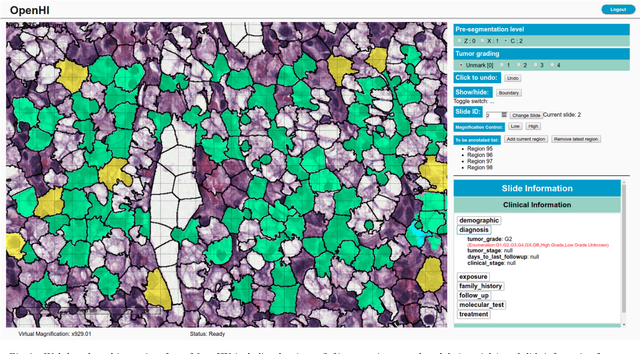
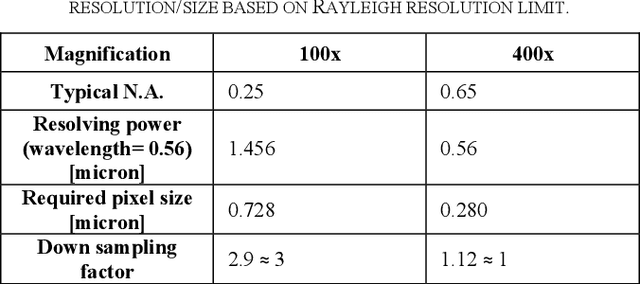
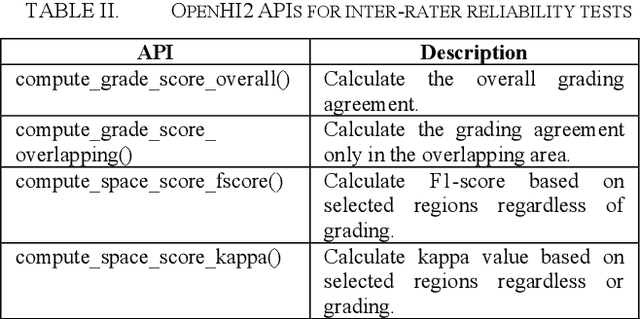
Abstract:Transition from conventional to digital pathology requires a new category of biomedical informatic infrastructure which could facilitate delicate pathological routine. Pathological diagnoses are sensitive to many external factors and is known to be subjective. Only systems that can meet strict requirements in pathology would be able to run along pathological routines and eventually digitized the study area, and the developed platform should comply with existing pathological routines and international standards. Currently, there are a number of available software tools which can perform histopathological tasks including virtual slide viewing, annotating, and basic image analysis, however, none of them can serve as a digital platform for pathology. Here we describe OpenHI2, an enhanced version Open Histopathological Image platform which is capable of supporting all basic pathological tasks and file formats; ready to be deployed in medical institutions on a standard server environment or cloud computing infrastructure. In this paper, we also describe the development decisions for the platform and propose solutions to overcome technical challenges so that OpenHI2 could be used as a platform for histopathological images. Further addition can be made to the platform since each component is modularized and fully documented. OpenHI2 is free, open-source, and available at https://gitlab.com/BioAI/OpenHI.
Effects of annotation granularity in deep learning models for histopathological images
Jan 14, 2020



Abstract:Pathological is crucial to cancer diagnosis. Usually, Pathologists draw their conclusion based on observed cell and tissue structure on histology slides. Rapid development in machine learning, especially deep learning have established robust and accurate classifiers. They are being used to analyze histopathological slides and assist pathologists in diagnosis. Most machine learning systems rely heavily on annotated data sets to gain experiences and knowledge to correctly and accurately perform various tasks such as classification and segmentation. This work investigates different granularity of annotations in histopathological data set including image-wise, bounding box, ellipse-wise, and pixel-wise to verify the influence of annotation in pathological slide on deep learning models. We design corresponding experiments to test classification and segmentation performance of deep learning models based on annotations with different annotation granularity. In classification, state-of-the-art deep learning-based classifiers perform better when trained by pixel-wise annotation dataset. On average, precision, recall and F1-score improves by 7.87%, 8.83% and 7.85% respectively. Thus, it is suggested that finer granularity annotations are better utilized by deep learning algorithms in classification tasks. Similarly, semantic segmentation algorithms can achieve 8.33% better segmentation accuracy when trained by pixel-wise annotations. Our study shows not only that finer-grained annotation can improve the performance of deep learning models, but also help extracts more accurate phenotypic information from histopathological slides. Intelligence systems trained on granular annotations may help pathologists inspecting certain regions for better diagnosis. The compartmentalized prediction approach similar to this work may contribute to phenotype and genotype association studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge