Xinrui Bao
Less is More for RAG: Information Gain Pruning for Generator-Aligned Reranking and Evidence Selection
Jan 24, 2026Abstract:Retrieval-augmented generation (RAG) grounds large language models with external evidence, but under a limited context budget, the key challenge is deciding which retrieved passages should be injected. We show that retrieval relevance metrics (e.g., NDCG) correlate weakly with end-to-end QA quality and can even become negatively correlated under multi-passage injection, where redundancy and mild conflicts destabilize generation. We propose \textbf{Information Gain Pruning (IGP)}, a deployment-friendly reranking-and-pruning module that selects evidence using a generator-aligned utility signal and filters weak or harmful passages before truncation, without changing existing budget interfaces. Across five open-domain QA benchmarks and multiple retrievers and generators, IGP consistently improves the quality--cost trade-off. In a representative multi-evidence setting, IGP delivers about +12--20% relative improvement in average F1 while reducing final-stage input tokens by roughly 76--79% compared to retriever-only baselines.
PIMIP: An Open Source Platform for Pathology Information Management and Integration
Nov 09, 2021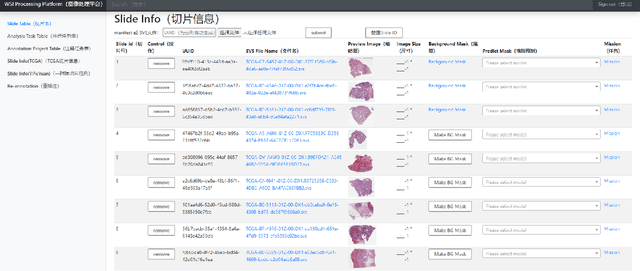
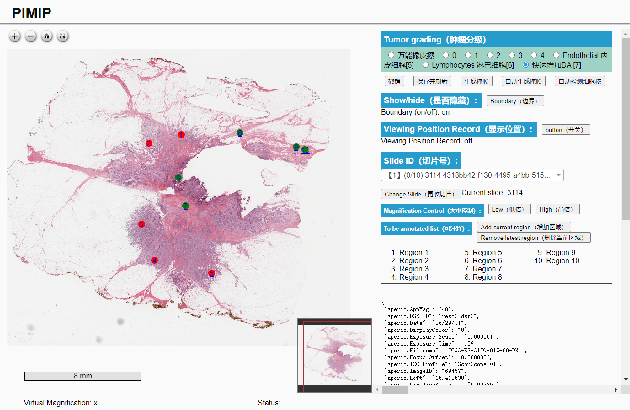
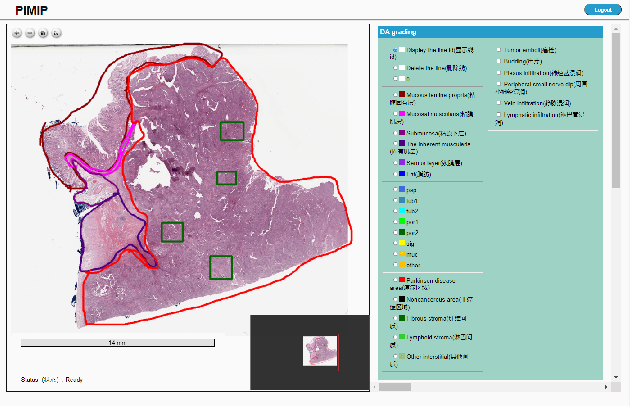
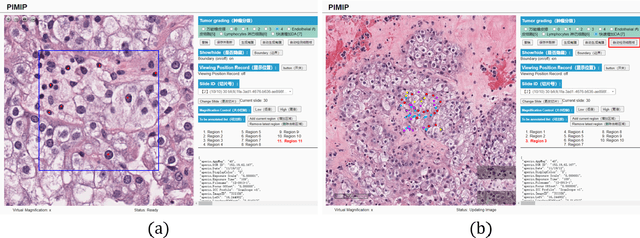
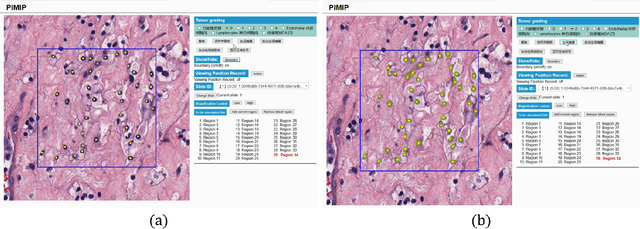
Abstract:Digital pathology plays a crucial role in the development of artificial intelligence in the medical field. The digital pathology platform can make the pathological resources digital and networked, and realize the permanent storage of visual data and the synchronous browsing processing without the limitation of time and space. It has been widely used in various fields of pathology. However, there is still a lack of an open and universal digital pathology platform to assist doctors in the management and analysis of digital pathological sections, as well as the management and structured description of relevant patient information. Most platforms cannot integrate image viewing, annotation and analysis, and text information management. To solve the above problems, we propose a comprehensive and extensible platform PIMIP. Our PIMIP has developed the image annotation functions based on the visualization of digital pathological sections. Our annotation functions support multi-user collaborative annotation and multi-device annotation, and realize the automation of some annotation tasks. In the annotation task, we invited a professional pathologist for guidance. We introduce a machine learning module for image analysis. The data we collected included public data from local hospitals and clinical examples. Our platform is more clinical and suitable for clinical use. In addition to image data, we also structured the management and display of text information. So our platform is comprehensive. The platform framework is built in a modular way to support users to add machine learning modules independently, which makes our platform extensible.
W-Net: A Two-Stage Convolutional Network for Nucleus Detection in Histopathology Image
Oct 26, 2021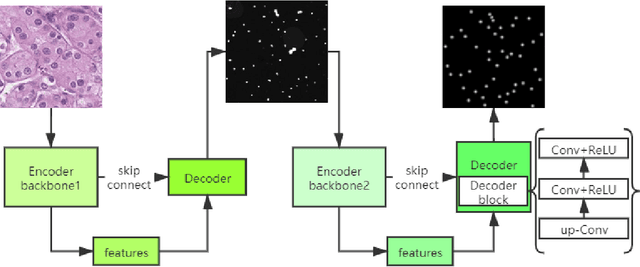
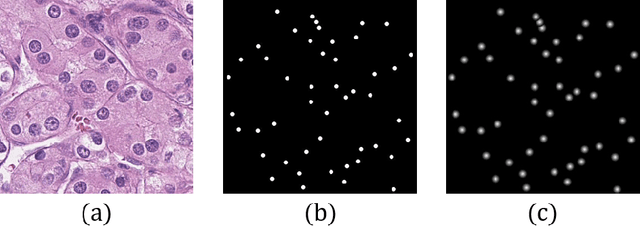
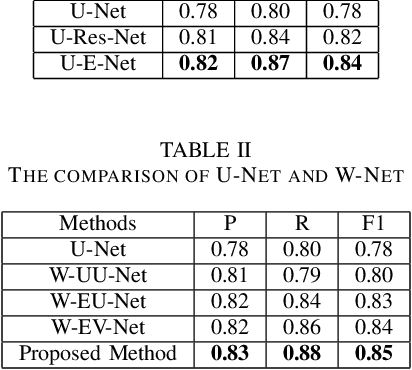
Abstract:Pathological diagnosis is the gold standard for cancer diagnosis, but it is labor-intensive, in which tasks such as cell detection, classification, and counting are particularly prominent. A common solution for automating these tasks is using nucleus segmentation technology. However, it is hard to train a robust nucleus segmentation model, due to several challenging problems, the nucleus adhesion, stacking, and excessive fusion with the background. Recently, some researchers proposed a series of automatic nucleus segmentation methods based on point annotation, which can significant improve the model performance. Nevertheless, the point annotation needs to be marked by experienced pathologists. In order to take advantage of segmentation methods based on point annotation, further alleviate the manual workload, and make cancer diagnosis more efficient and accurate, it is necessary to develop an automatic nucleus detection algorithm, which can automatically and efficiently locate the position of the nucleus in the pathological image and extract valuable information for pathologists. In this paper, we propose a W-shaped network for automatic nucleus detection. Different from the traditional U-Net based method, mapping the original pathology image to the target mask directly, our proposed method split the detection task into two sub-tasks. The first sub-task maps the original pathology image to the binary mask, then the binary mask is mapped to the density mask in the second sub-task. After the task is split, the task's difficulty is significantly reduced, and the network's overall performance is improved.
A Precision Diagnostic Framework of Renal Cell Carcinoma on Whole-Slide Images using Deep Learning
Oct 26, 2021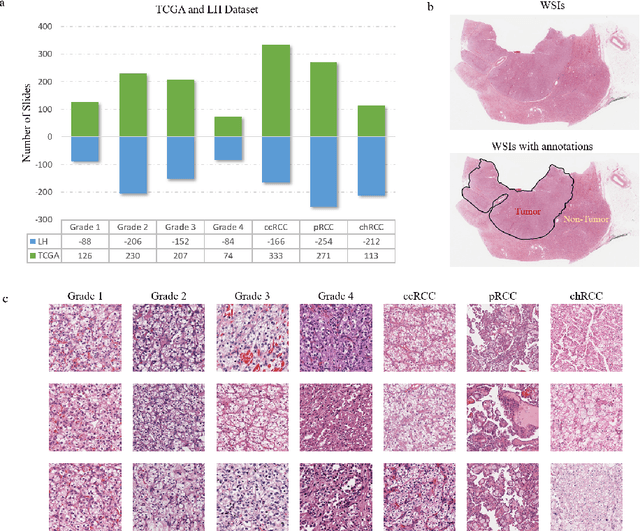
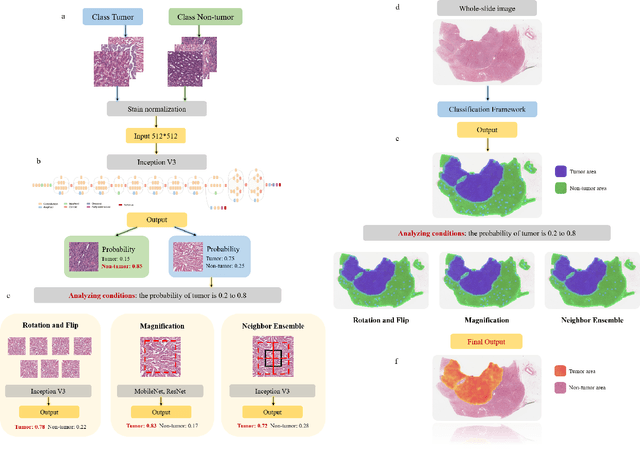
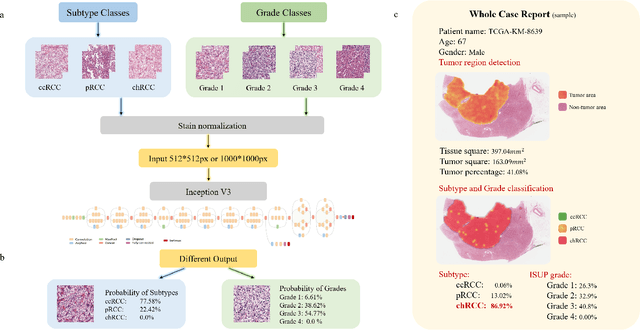
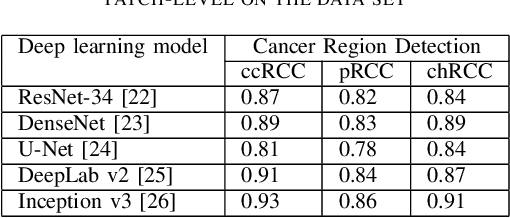
Abstract:Diagnostic pathology, which is the basis and gold standard of cancer diagnosis, provides essential information on the prognosis of the disease and vital evidence for clinical treatment. Tumor region detection, subtype and grade classification are the fundamental diagnostic indicators for renal cell carcinoma (RCC) in whole-slide images (WSIs). However, pathological diagnosis is subjective, differences in observation and diagnosis between pathologists is common in hospitals with inadequate diagnostic capacity. The main challenge for developing deep learning based RCC diagnostic system is the lack of large-scale datasets with precise annotations. In this work, we proposed a deep learning-based framework for analyzing histopathological images of patients with renal cell carcinoma, which has the potential to achieve pathologist-level accuracy in diagnosis. A deep convolutional neural network (InceptionV3) was trained on the high-quality annotated dataset of The Cancer Genome Atlas (TCGA) whole-slide histopathological image for accurate tumor area detection, classification of RCC subtypes, and ISUP grades classification of clear cell carcinoma subtypes. These results suggest that our framework can help pathologists in the detection of cancer region and classification of subtypes and grades, which could be applied to any cancer type, providing auxiliary diagnosis and promoting clinical consensus.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge