Guillaume Richard
Multi-modal Transfer Learning between Biological Foundation Models
Jun 20, 2024



Abstract:Biological sequences encode fundamental instructions for the building blocks of life, in the form of DNA, RNA, and proteins. Modeling these sequences is key to understand disease mechanisms and is an active research area in computational biology. Recently, Large Language Models have shown great promise in solving certain biological tasks but current approaches are limited to a single sequence modality (DNA, RNA, or protein). Key problems in genomics intrinsically involve multiple modalities, but it remains unclear how to adapt general-purpose sequence models to those cases. In this work we propose a multi-modal model that connects DNA, RNA, and proteins by leveraging information from different pre-trained modality-specific encoders. We demonstrate its capabilities by applying it to the largely unsolved problem of predicting how multiple RNA transcript isoforms originate from the same gene (i.e. same DNA sequence) and map to different transcription expression levels across various human tissues. We show that our model, dubbed IsoFormer, is able to accurately predict differential transcript expression, outperforming existing methods and leveraging the use of multiple modalities. Our framework also achieves efficient transfer knowledge from the encoders pre-training as well as in between modalities. We open-source our model, paving the way for new multi-modal gene expression approaches.
PASTA: Pretrained Action-State Transformer Agents
Jul 20, 2023



Abstract:Self-supervised learning has brought about a revolutionary paradigm shift in various computing domains, including NLP, vision, and biology. Recent approaches involve pre-training transformer models on vast amounts of unlabeled data, serving as a starting point for efficiently solving downstream tasks. In the realm of reinforcement learning, researchers have recently adapted these approaches by developing models pre-trained on expert trajectories, enabling them to address a wide range of tasks, from robotics to recommendation systems. However, existing methods mostly rely on intricate pre-training objectives tailored to specific downstream applications. This paper presents a comprehensive investigation of models we refer to as Pretrained Action-State Transformer Agents (PASTA). Our study uses a unified methodology and covers an extensive set of general downstream tasks including behavioral cloning, offline RL, sensor failure robustness, and dynamics change adaptation. Our goal is to systematically compare various design choices and provide valuable insights to practitioners for building robust models. Key highlights of our study include tokenization at the action and state component level, using fundamental pre-training objectives like next token prediction, training models across diverse domains simultaneously, and using parameter efficient fine-tuning (PEFT). The developed models in our study contain fewer than 10 million parameters and the application of PEFT enables fine-tuning of fewer than 10,000 parameters during downstream adaptation, allowing a broad community to use these models and reproduce our experiments. We hope that this study will encourage further research into the use of transformers with first-principles design choices to represent RL trajectories and contribute to robust policy learning.
Gradient-Informed Quality Diversity for the Illumination of Discrete Spaces
Jun 08, 2023Abstract:Quality Diversity (QD) algorithms have been proposed to search for a large collection of both diverse and high-performing solutions instead of a single set of local optima. While early QD algorithms view the objective and descriptor functions as black-box functions, novel tools have been introduced to use gradient information to accelerate the search and improve overall performance of those algorithms over continuous input spaces. However a broad range of applications involve discrete spaces, such as drug discovery or image generation. Exploring those spaces is challenging as they are combinatorially large and gradients cannot be used in the same manner as in continuous spaces. We introduce map-elites with a Gradient-Informed Discrete Emitter (ME-GIDE), which extends QD optimisation with differentiable functions over discrete search spaces. ME-GIDE leverages the gradient information of the objective and descriptor functions with respect to its discrete inputs to propose gradient-informed updates that guide the search towards a diverse set of high quality solutions. We evaluate our method on challenging benchmarks including protein design and discrete latent space illumination and find that our method outperforms state-of-the-art QD algorithms in all benchmarks.
The Quality-Diversity Transformer: Generating Behavior-Conditioned Trajectories with Decision Transformers
Mar 27, 2023Abstract:In the context of neuroevolution, Quality-Diversity algorithms have proven effective in generating repertoires of diverse and efficient policies by relying on the definition of a behavior space. A natural goal induced by the creation of such a repertoire is trying to achieve behaviors on demand, which can be done by running the corresponding policy from the repertoire. However, in uncertain environments, two problems arise. First, policies can lack robustness and repeatability, meaning that multiple episodes under slightly different conditions often result in very different behaviors. Second, due to the discrete nature of the repertoire, solutions vary discontinuously. Here we present a new approach to achieve behavior-conditioned trajectory generation based on two mechanisms: First, MAP-Elites Low-Spread (ME-LS), which constrains the selection of solutions to those that are the most consistent in the behavior space. Second, the Quality-Diversity Transformer (QDT), a Transformer-based model conditioned on continuous behavior descriptors, which trains on a dataset generated by policies from a ME-LS repertoire and learns to autoregressively generate sequences of actions that achieve target behaviors. Results show that ME-LS produces consistent and robust policies, and that its combination with the QDT yields a single policy capable of achieving diverse behaviors on demand with high accuracy.
Multi-Objective Quality Diversity Optimization
Feb 07, 2022



Abstract:In this work, we consider the problem of Quality-Diversity (QD) optimization with multiple objectives. QD algorithms have been proposed to search for a large collection of both diverse and high-performing solutions instead of a single set of local optima. Thriving for diversity was shown to be useful in many industrial and robotics applications. On the other hand, most real-life problems exhibit several potentially antagonist objectives to be optimized. Hence being able to optimize for multiple objectives with an appropriate technique while thriving for diversity is important to many fields. Here, we propose an extension of the MAP-Elites algorithm in the multi-objective setting: Multi-Objective MAP-Elites (MOME). Namely, it combines the diversity inherited from the MAP-Elites grid algorithm with the strength of multi-objective optimizations by filling each cell with a Pareto Front. As such, it allows to extract diverse solutions in the descriptor space while exploring different compromises between objectives. We evaluate our method on several tasks, from standard optimization problems to robotics simulations. Our experimental evaluation shows the ability of MOME to provide diverse solutions while providing global performances similar to standard multi-objective algorithms.
Unsupervised Domain Adaptation for Constraining Star Formation Histories
Dec 28, 2021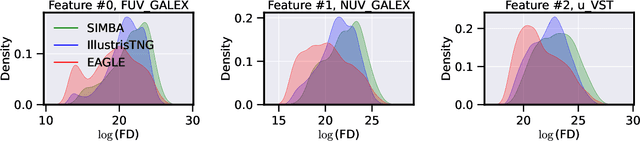
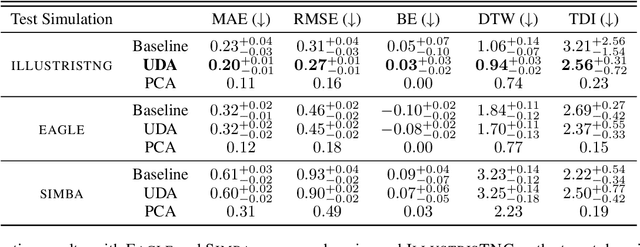
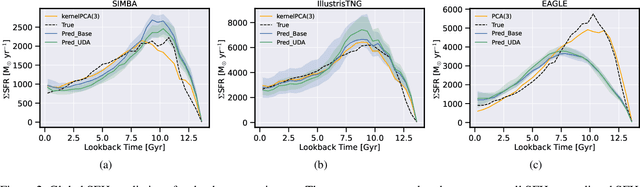
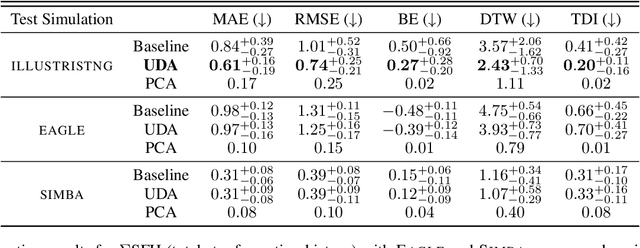
Abstract:The prevalent paradigm of machine learning today is to use past observations to predict future ones. What if, however, we are interested in knowing the past given the present? This situation is indeed one that astronomers must contend with often. To understand the formation of our universe, we must derive the time evolution of the visible mass content of galaxies. However, to observe a complete star life, one would need to wait for one billion years! To overcome this difficulty, astrophysicists leverage supercomputers and evolve simulated models of galaxies till the current age of the universe, thus establishing a mapping between observed radiation and star formation histories (SFHs). Such ground-truth SFHs are lacking for actual galaxy observations, where they are usually inferred -- with often poor confidence -- from spectral energy distributions (SEDs) using Bayesian fitting methods. In this investigation, we discuss the ability of unsupervised domain adaptation to derive accurate SFHs for galaxies with simulated data as a necessary first step in developing a technique that can ultimately be applied to observational data.
ADAPT : Awesome Domain Adaptation Python Toolbox
Jul 07, 2021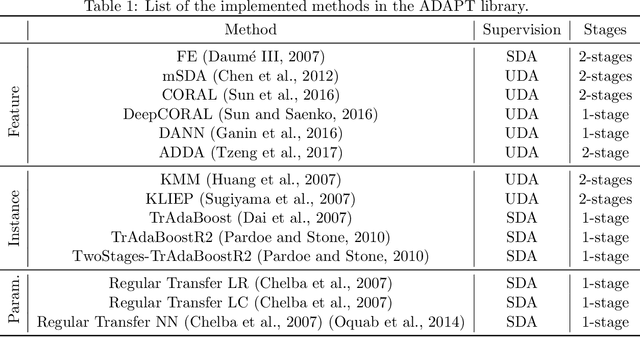
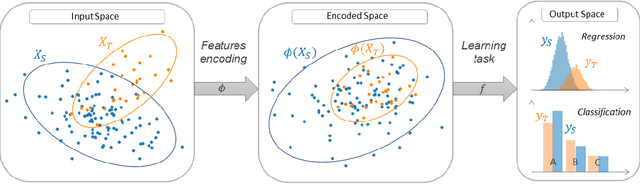
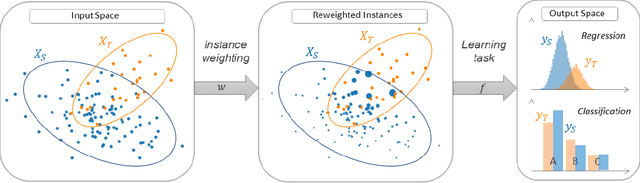
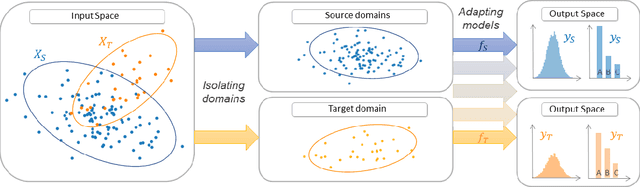
Abstract:ADAPT is an open-source python library providing the implementation of several domain adaptation methods. The library is suited for scikit-learn estimator object (object which implement fit and predict methods) and tensorflow models. Most of the implemented methods are developed in an estimator agnostic fashion, offering various possibilities adapted to multiple usage. The library offers three modules corresponding to the three principal strategies of domain adaptation: (i) feature-based containing methods performing feature transformation; (ii) instance-based with the implementation of reweighting techniques and (iii) parameter-based proposing methods to adapt pre-trained models to novel observations. A full documentation is proposed online https://adapt-python.github.io/adapt/ with gallery of examples. Besides, the library presents an high test coverage.
Adversarial Weighting for Domain Adaptation in Regression
Jun 15, 2020



Abstract:We present a novel instance based approach to handle regression tasks in the context of supervised domain adaptation. The approach developed in this paper relies on the assumption that the task on the target domain can be efficiently learned by adequately reweighting the source instances during training phase. We introduce a novel formulation of the optimization objective for domain adaptation which relies on a discrepancy distance characterizing the difference between domains according to a specific task and a class of hypotheses. To solve this problem, we develop an adversarial network algorithm which learns both the source weighting scheme and the task in one feed-forward gradient descent. We provide numerical evidence of the relevance of the method on public datasets for domain adaptation through reproducible experiments accessible via an online demo interface.
Autoencoder-based time series clustering with energy applications
Feb 10, 2020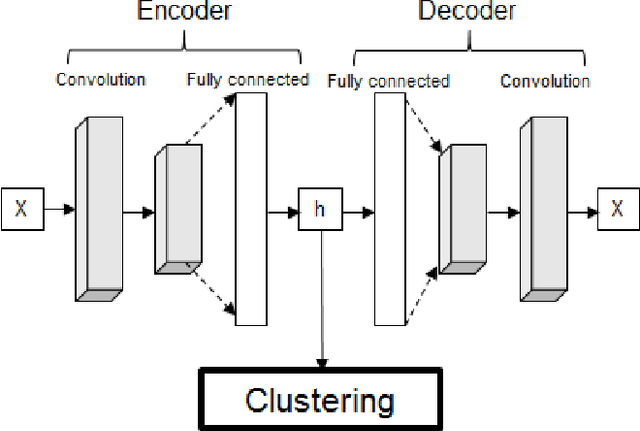
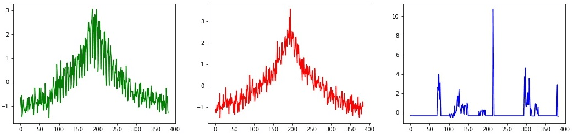
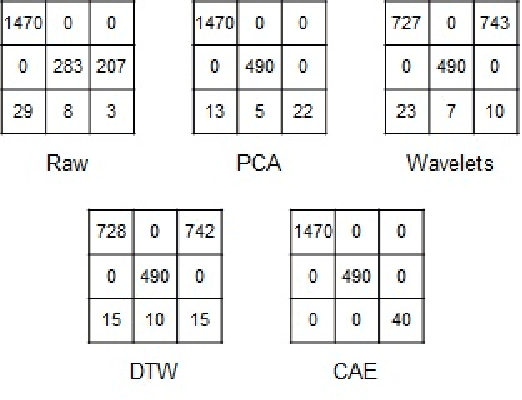
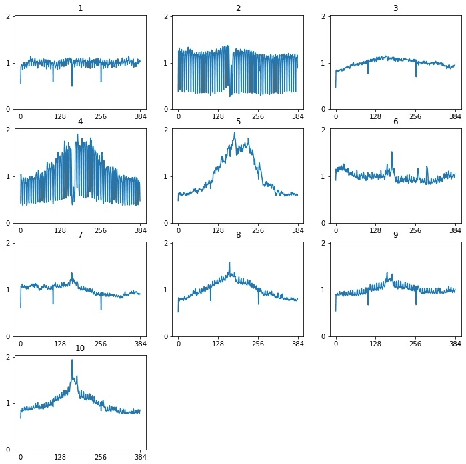
Abstract:Time series clustering is a challenging task due to the specific nature of the data. Classical approaches do not perform well and need to be adapted either through a new distance measure or a data transformation. In this paper we investigate the combination of a convolutional autoencoder and a k-medoids algorithm to perfom time series clustering. The convolutional autoencoder allows to extract meaningful features and reduce the dimension of the data, leading to an improvement of the subsequent clustering. Using simulation and energy related data to validate the approach, experimental results show that the clustering is robust to outliers thus leading to finer clusters than with standard methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge