Bernardo P. de Almeida
Simple Guidance Mechanisms for Discrete Diffusion Models
Dec 13, 2024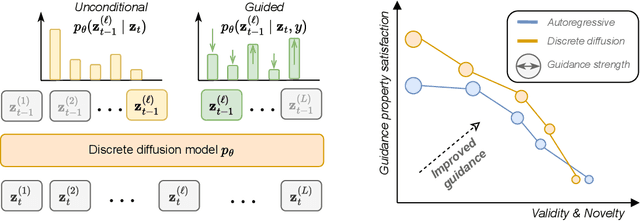
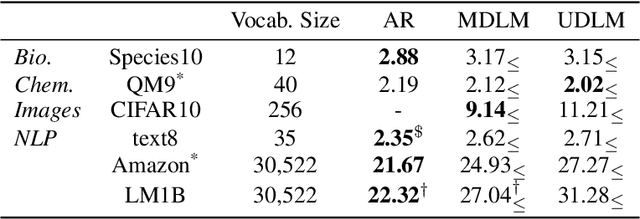
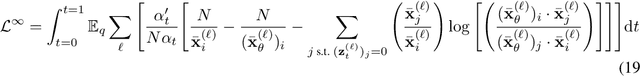
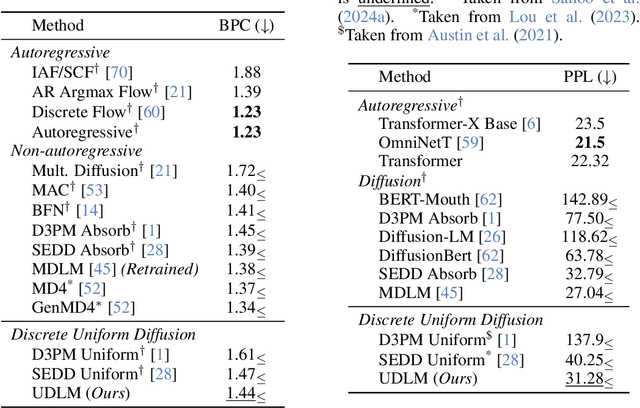
Abstract:Diffusion models for continuous data gained widespread adoption owing to their high quality generation and control mechanisms. However, controllable diffusion on discrete data faces challenges given that continuous guidance methods do not directly apply to discrete diffusion. Here, we provide a straightforward derivation of classifier-free and classifier-based guidance for discrete diffusion, as well as a new class of diffusion models that leverage uniform noise and that are more guidable because they can continuously edit their outputs. We improve the quality of these models with a novel continuous-time variational lower bound that yields state-of-the-art performance, especially in settings involving guidance or fast generation. Empirically, we demonstrate that our guidance mechanisms combined with uniform noise diffusion improve controllable generation relative to autoregressive and diffusion baselines on several discrete data domains, including genomic sequences, small molecule design, and discretized image generation.
Multi-modal Transfer Learning between Biological Foundation Models
Jun 20, 2024



Abstract:Biological sequences encode fundamental instructions for the building blocks of life, in the form of DNA, RNA, and proteins. Modeling these sequences is key to understand disease mechanisms and is an active research area in computational biology. Recently, Large Language Models have shown great promise in solving certain biological tasks but current approaches are limited to a single sequence modality (DNA, RNA, or protein). Key problems in genomics intrinsically involve multiple modalities, but it remains unclear how to adapt general-purpose sequence models to those cases. In this work we propose a multi-modal model that connects DNA, RNA, and proteins by leveraging information from different pre-trained modality-specific encoders. We demonstrate its capabilities by applying it to the largely unsolved problem of predicting how multiple RNA transcript isoforms originate from the same gene (i.e. same DNA sequence) and map to different transcription expression levels across various human tissues. We show that our model, dubbed IsoFormer, is able to accurately predict differential transcript expression, outperforming existing methods and leveraging the use of multiple modalities. Our framework also achieves efficient transfer knowledge from the encoders pre-training as well as in between modalities. We open-source our model, paving the way for new multi-modal gene expression approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge