George H. Chen
Deep Kernel Aalen-Johansen Estimator: An Interpretable and Flexible Neural Net Framework for Competing Risks
Dec 08, 2025Abstract:We propose an interpretable deep competing risks model called the Deep Kernel Aalen-Johansen (DKAJ) estimator, which generalizes the classical Aalen-Johansen nonparametric estimate of cumulative incidence functions (CIFs). Each data point (e.g., patient) is represented as a weighted combination of clusters. If a data point has nonzero weight only for one cluster, then its predicted CIFs correspond to those of the classical Aalen-Johansen estimator restricted to data points from that cluster. These weights come from an automatically learned kernel function that measures how similar any two data points are. On four standard competing risks datasets, we show that DKAJ is competitive with state-of-the-art baselines while being able to provide visualizations to assist model interpretation.
Explaining the Success of Nearest Neighbor Methods in Prediction
Feb 21, 2025Abstract:Many modern methods for prediction leverage nearest neighbor search to find past training examples most similar to a test example, an idea that dates back in text to at least the 11th century and has stood the test of time. This monograph aims to explain the success of these methods, both in theory, for which we cover foundational nonasymptotic statistical guarantees on nearest-neighbor-based regression and classification, and in practice, for which we gather prominent methods for approximate nearest neighbor search that have been essential to scaling prediction systems reliant on nearest neighbor analysis to handle massive datasets. Furthermore, we discuss connections to learning distances for use with nearest neighbor methods, including how random decision trees and ensemble methods learn nearest neighbor structure, as well as recent developments in crowdsourcing and graphons. In terms of theory, our focus is on nonasymptotic statistical guarantees, which we state in the form of how many training data and what algorithm parameters ensure that a nearest neighbor prediction method achieves a user-specified error tolerance. We begin with the most general of such results for nearest neighbor and related kernel regression and classification in general metric spaces. In such settings in which we assume very little structure, what enables successful prediction is smoothness in the function being estimated for regression, and a low probability of landing near the decision boundary for classification. In practice, these conditions could be difficult to verify for a real dataset. We then cover recent guarantees on nearest neighbor prediction in the three case studies of time series forecasting, recommending products to people over time, and delineating human organs in medical images by looking at image patches. In these case studies, clustering structure enables successful prediction.
Generalized Prompt Tuning: Adapting Frozen Univariate Time Series Foundation Models for Multivariate Healthcare Time Series
Nov 19, 2024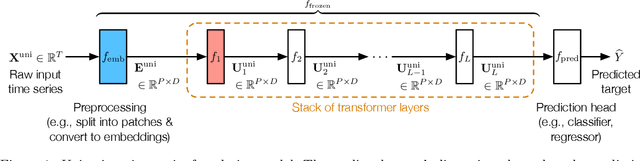
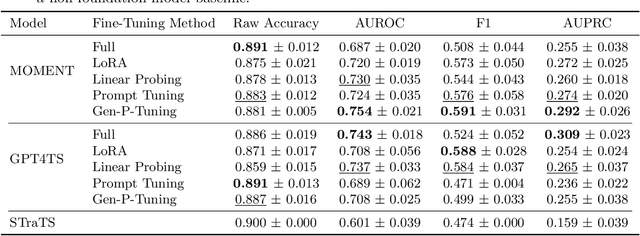
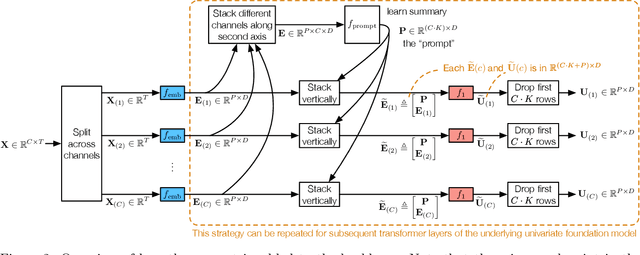
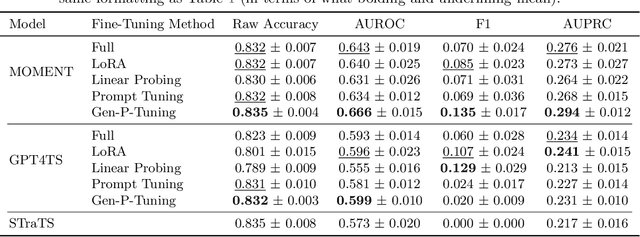
Abstract:Time series foundation models are pre-trained on large datasets and are able to achieve state-of-the-art performance in diverse tasks. However, to date, there has been limited work demonstrating how well these models perform in medical applications, where labeled data can be scarce. Further, we observe that currently, the majority of time series foundation models either are univariate in nature, or assume channel independence, meaning that they handle multivariate time series but do not model how the different variables relate. In this paper, we propose a prompt-tuning-inspired fine-tuning technique, Generalized Prompt Tuning (Gen-P-Tuning), that enables us to adapt an existing univariate time series foundation model (treated as frozen) to handle multivariate time series prediction. Our approach provides a way to combine information across channels (variables) of multivariate time series. We demonstrate the effectiveness of our fine-tuning approach against various baselines on two MIMIC classification tasks, and on influenza-like illness forecasting.
An Introduction to Deep Survival Analysis Models for Predicting Time-to-Event Outcomes
Oct 01, 2024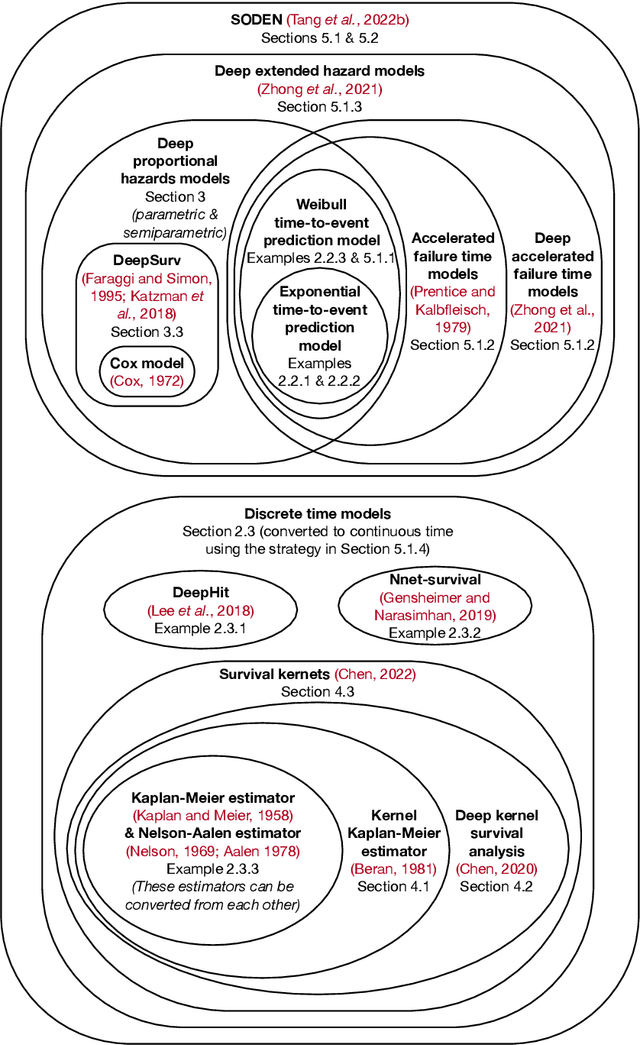
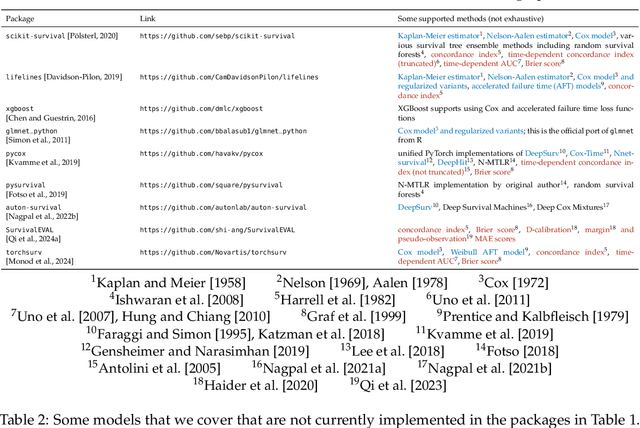
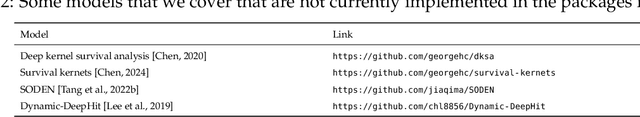

Abstract:Many applications involve reasoning about time durations before a critical event happens--also called time-to-event outcomes. When will a customer cancel a subscription, a coma patient wake up, or a convicted criminal reoffend? Time-to-event outcomes have been studied extensively within the field of survival analysis primarily by the statistical, medical, and reliability engineering communities, with textbooks already available in the 1970s and '80s. This monograph aims to provide a reasonably self-contained modern introduction to survival analysis. We focus on predicting time-to-event outcomes at the individual data point level with the help of neural networks. Our goal is to provide the reader with a working understanding of precisely what the basic time-to-event prediction problem is, how it differs from standard regression and classification, and how key "design patterns" have been used time after time to derive new time-to-event prediction models, from classical methods like the Cox proportional hazards model to modern deep learning approaches such as deep kernel Kaplan-Meier estimators and neural ordinary differential equation models. We further delve into two extensions of the basic time-to-event prediction setup: predicting which of several critical events will happen first along with the time until this earliest event happens (the competing risks setting), and predicting time-to-event outcomes given a time series that grows in length over time (the dynamic setting). We conclude with a discussion of a variety of topics such as fairness, causal reasoning, interpretability, and statistical guarantees. Our monograph comes with an accompanying code repository that implements every model and evaluation metric that we cover in detail.
Temporal Supervised Contrastive Learning for Modeling Patient Risk Progression
Dec 10, 2023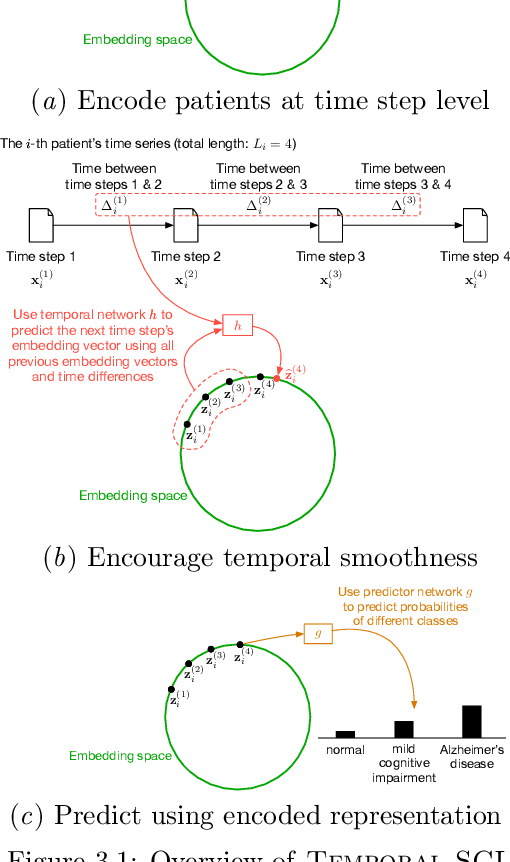
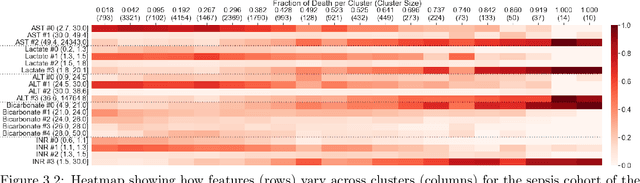
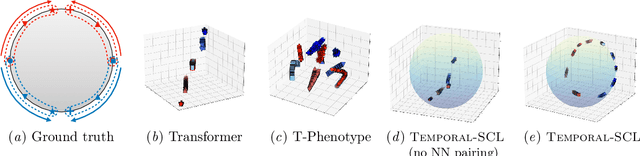
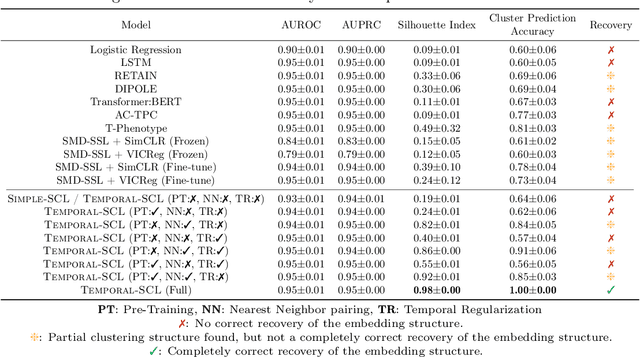
Abstract:We consider the problem of predicting how the likelihood of an outcome of interest for a patient changes over time as we observe more of the patient data. To solve this problem, we propose a supervised contrastive learning framework that learns an embedding representation for each time step of a patient time series. Our framework learns the embedding space to have the following properties: (1) nearby points in the embedding space have similar predicted class probabilities, (2) adjacent time steps of the same time series map to nearby points in the embedding space, and (3) time steps with very different raw feature vectors map to far apart regions of the embedding space. To achieve property (3), we employ a nearest neighbor pairing mechanism in the raw feature space. This mechanism also serves as an alternative to data augmentation, a key ingredient of contrastive learning, which lacks a standard procedure that is adequately realistic for clinical tabular data, to our knowledge. We demonstrate that our approach outperforms state-of-the-art baselines in predicting mortality of septic patients (MIMIC-III dataset) and tracking progression of cognitive impairment (ADNI dataset). Our method also consistently recovers the correct synthetic dataset embedding structure across experiments, a feat not achieved by baselines. Our ablation experiments show the pivotal role of our nearest neighbor pairing.
Neurological Prognostication of Post-Cardiac-Arrest Coma Patients Using EEG Data: A Dynamic Survival Analysis Framework with Competing Risks
Aug 17, 2023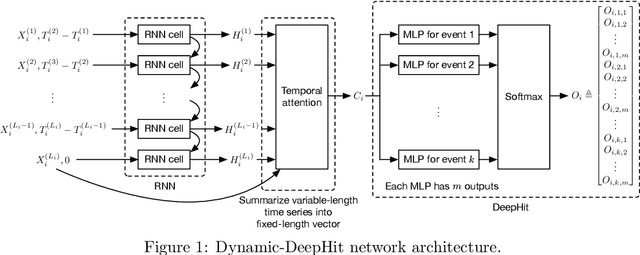
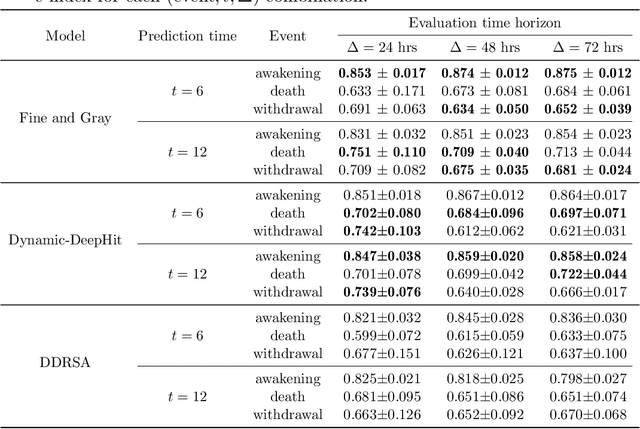
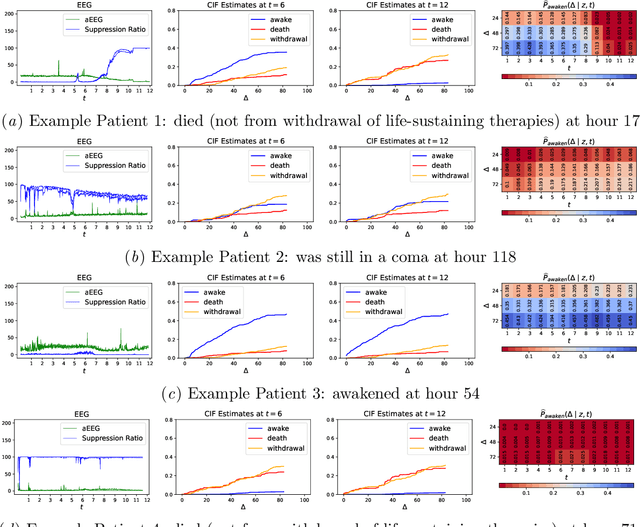

Abstract:Patients resuscitated from cardiac arrest who enter a coma are at high risk of death. Forecasting neurological outcomes of these patients (the task of neurological prognostication) could help with treatment decisions. In this paper, we propose, to the best of our knowledge, the first dynamic framework for neurological prognostication of post-cardiac-arrest comatose patients using EEG data: our framework makes predictions for a patient over time as more EEG data become available, and different training patients' available EEG time series could vary in length. Predictions are phrased in terms of either time-to-event outcomes (time-to-awakening or time-to-death) or as the patient's probability of awakening or of dying across multiple time horizons. Our framework uses any dynamic survival analysis model that supports competing risks in the form of estimating patient-level cumulative incidence functions. We consider three competing risks as to what happens first to a patient: awakening, being withdrawn from life-sustaining therapies (and thus deterministically dying), or dying (by other causes). We demonstrate our framework by benchmarking three existing dynamic survival analysis models that support competing risks on a real dataset of 922 patients. Our main experimental findings are that: (1) the classical Fine and Gray model which only uses a patient's static features and summary statistics from the patient's latest hour's worth of EEG data is highly competitive, achieving accuracy scores as high as the recently developed Dynamic-DeepHit model that uses substantially more of the patient's EEG data; and (2) in an ablation study, we show that our choice of modeling three competing risks results in a model that is at least as accurate while learning more information than simpler models (using two competing risks or a standard survival analysis setup with no competing risks).
Improving Fairness in Deepfake Detection
Jun 29, 2023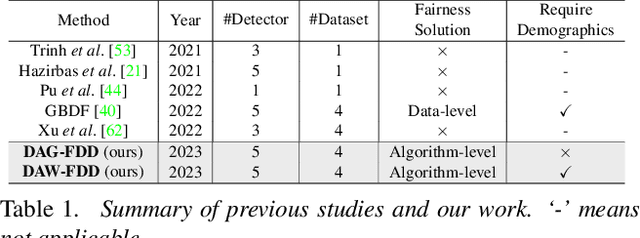
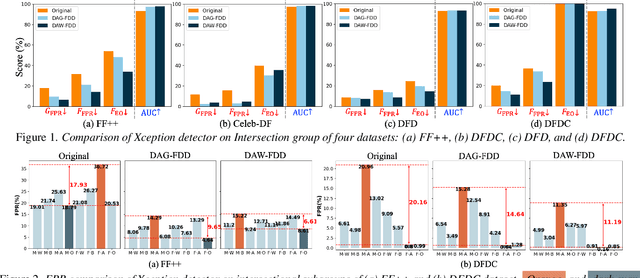
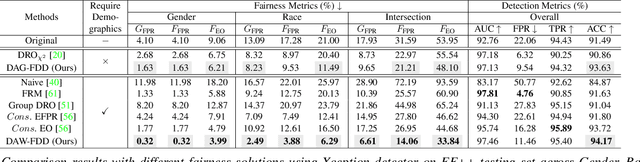
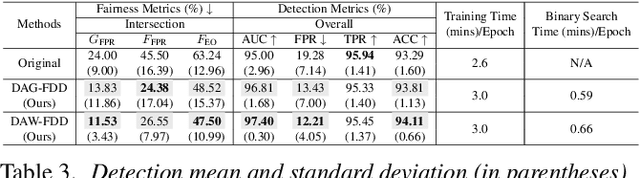
Abstract:Despite the development of effective deepfake detection models in recent years, several recent studies have demonstrated that biases in the training data utilized to develop deepfake detection models can lead to unfair performance for demographic groups of different races and/or genders. Such can result in these groups being unfairly targeted or excluded from detection, allowing misclassified deepfakes to manipulate public opinion and erode trust in the model. While these studies have focused on identifying and evaluating the unfairness in deepfake detection, no methods have been developed to address the fairness issue of deepfake detection at the algorithm level. In this work, we make the first attempt to improve deepfake detection fairness by proposing novel loss functions to train fair deepfake detection models in ways that are agnostic or aware of demographic factors. Extensive experiments on four deepfake datasets and five deepfake detectors demonstrate the effectiveness and flexibility of our approach in improving the deepfake detection fairness.
A General Framework for Visualizing Embedding Spaces of Neural Survival Analysis Models Based on Angular Information
May 11, 2023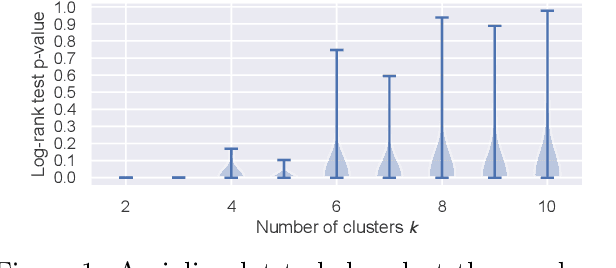

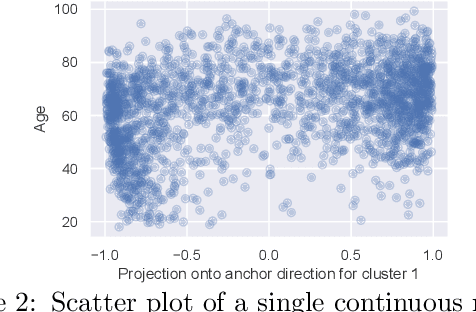
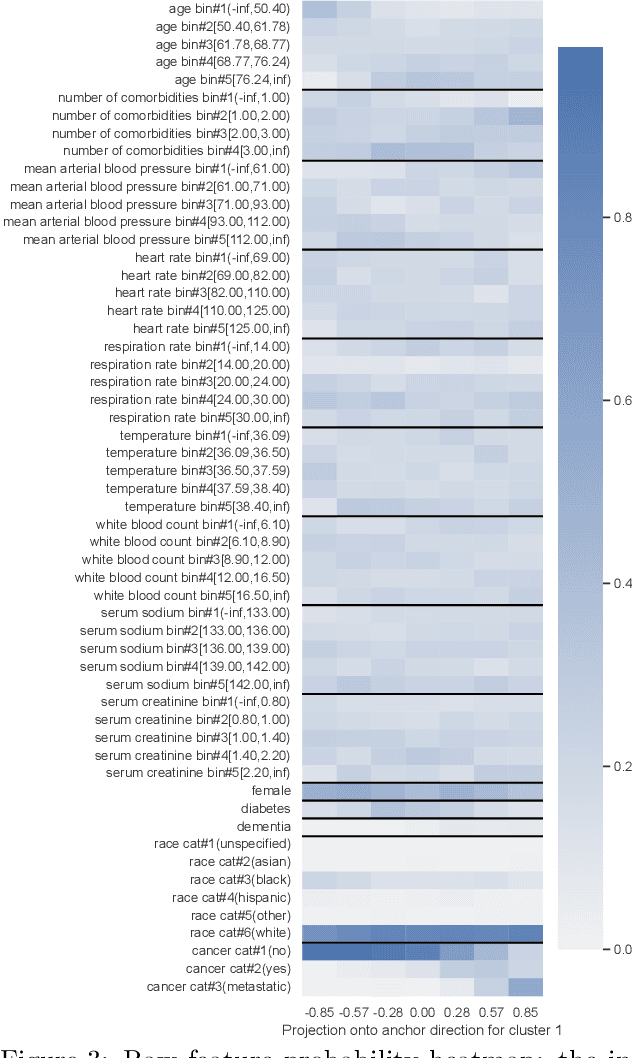
Abstract:We propose a general framework for visualizing any intermediate embedding representation used by any neural survival analysis model. Our framework is based on so-called anchor directions in an embedding space. We show how to estimate these anchor directions using clustering or, alternatively, using user-supplied "concepts" defined by collections of raw inputs (e.g., feature vectors all from female patients could encode the concept "female"). For tabular data, we present visualization strategies that reveal how anchor directions relate to raw clinical features and to survival time distributions. We then show how these visualization ideas extend to handling raw inputs that are images. Our framework is built on looking at angles between vectors in an embedding space, where there could be "information loss" by ignoring magnitude information. We show how this loss results in a "clumping" artifact that appears in our visualizations, and how to reduce this information loss in practice.
Distributionally Robust Survival Analysis: A Novel Fairness Loss Without Demographics
Nov 18, 2022Abstract:We propose a general approach for training survival analysis models that minimizes a worst-case error across all subpopulations that are large enough (occurring with at least a user-specified minimum probability). This approach uses a training loss function that does not know any demographic information to treat as sensitive. Despite this, we demonstrate that our proposed approach often scores better on recently established fairness metrics (without a significant drop in prediction accuracy) compared to various baselines, including ones which directly use sensitive demographic information in their training loss. Our code is available at: https://github.com/discovershu/DRO_COX
Survival Kernets: Scalable and Interpretable Deep Kernel Survival Analysis with an Accuracy Guarantee
Jun 30, 2022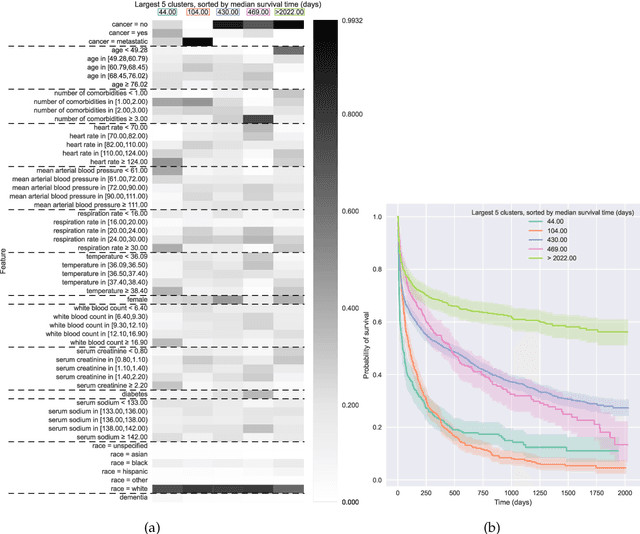
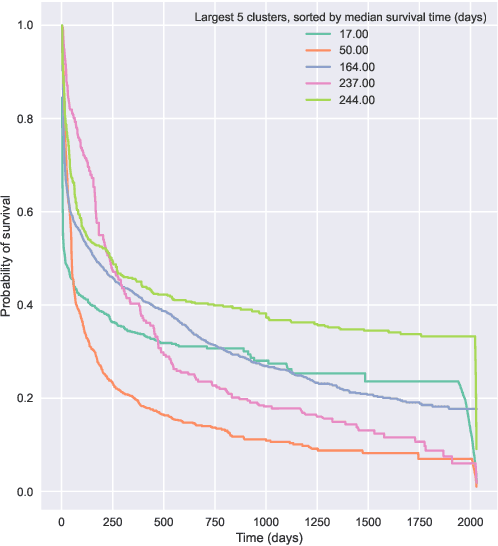
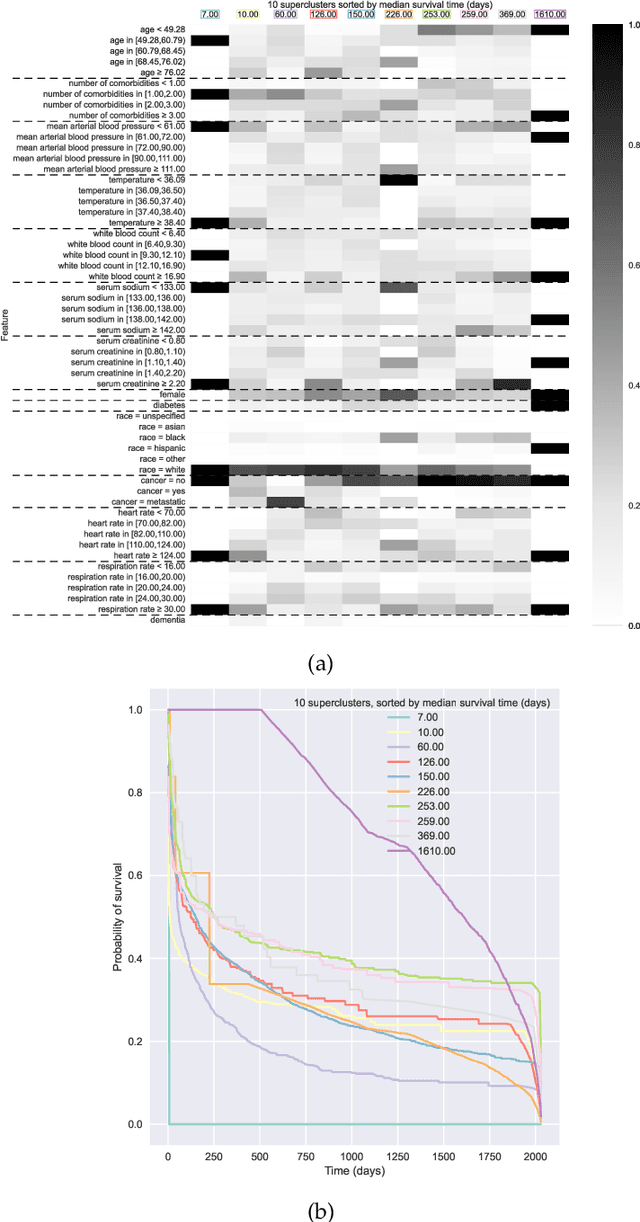
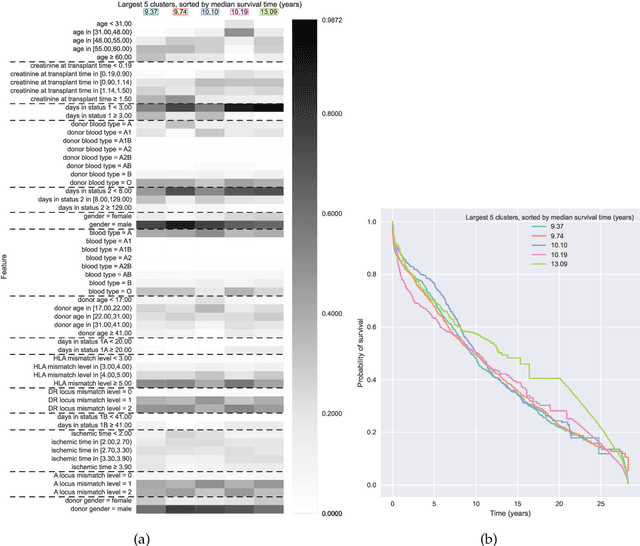
Abstract:Kernel survival analysis models estimate individual survival distributions with the help of a kernel function, which measures the similarity between any two data points. Such a kernel function can be learned using deep kernel survival models. In this paper, we present a new deep kernel survival model called a survival kernet, which scales to large datasets in a manner that is amenable to model interpretation and also theoretical analysis. Specifically, the training data are partitioned into clusters based on a recently developed training set compression scheme for classification and regression called kernel netting that we extend to the survival analysis setting. At test-time, each data point is represented as a weighted combination of these clusters, and each such cluster can be visualized. For a special case of survival kernets, we establish a finite-sample error bound on predicted survival distributions that is, up to a log factor, optimal. Whereas scalability at test time is achieved using the aforementioned kernel netting compression strategy, scalability during training is achieved by a warm-start procedure based on tree ensembles such as XGBoost and a heuristic approach to accelerating neural architecture search. On three standard survival analysis datasets of varying sizes (up to roughly 3 million data points), we show that survival kernets are highly competitive with the best of baselines tested in terms of concordance index. Our code is available at: https://github.com/georgehc/survival-kernets
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge