Jeremy C. Weiss
MIMIC-\RNum{4}-Ext-22MCTS: A 22 Millions-Event Temporal Clinical Time-Series Dataset with Relative Timestamp for Risk Prediction
May 01, 2025



Abstract:Clinical risk prediction based on machine learning algorithms plays a vital role in modern healthcare. A crucial component in developing a reliable prediction model is collecting high-quality time series clinical events. In this work, we release such a dataset that consists of 22,588,586 Clinical Time Series events, which we term MIMIC-\RNum{4}-Ext-22MCTS. Our source data are discharge summaries selected from the well-known yet unstructured MIMIC-IV-Note \cite{Johnson2023-pg}. We then extract clinical events as short text span from the discharge summaries, along with the timestamps of these events as temporal information. The general-purpose MIMIC-IV-Note pose specific challenges for our work: it turns out that the discharge summaries are too lengthy for typical natural language models to process, and the clinical events of interest often are not accompanied with explicit timestamps. Therefore, we propose a new framework that works as follows: 1) we break each discharge summary into manageably small text chunks; 2) we apply contextual BM25 and contextual semantic search to retrieve chunks that have a high potential of containing clinical events; and 3) we carefully design prompts to teach the recently released Llama-3.1-8B \cite{touvron2023llama} model to identify or infer temporal information of the chunks. We show that the obtained dataset is so informative and transparent that standard models fine-tuned on our dataset are achieving significant improvements in healthcare applications. In particular, the BERT model fine-tuned based on our dataset achieves 10\% improvement in accuracy on medical question answering task, and 3\% improvement in clinical trial matching task compared with the classic BERT. The GPT-2 model, fine-tuned on our dataset, produces more clinically reliable results for clinical questions.
Forecasting from Clinical Textual Time Series: Adaptations of the Encoder and Decoder Language Model Families
Apr 14, 2025Abstract:Clinical case reports encode rich, temporal patient trajectories that are often underexploited by traditional machine learning methods relying on structured data. In this work, we introduce the forecasting problem from textual time series, where timestamped clinical findings--extracted via an LLM-assisted annotation pipeline--serve as the primary input for prediction. We systematically evaluate a diverse suite of models, including fine-tuned decoder-based large language models and encoder-based transformers, on tasks of event occurrence prediction, temporal ordering, and survival analysis. Our experiments reveal that encoder-based models consistently achieve higher F1 scores and superior temporal concordance for short- and long-horizon event forecasting, while fine-tuned masking approaches enhance ranking performance. In contrast, instruction-tuned decoder models demonstrate a relative advantage in survival analysis, especially in early prognosis settings. Our sensitivity analyses further demonstrate the importance of time ordering, which requires clinical time series construction, as compared to text ordering, the format of the text inputs that LLMs are classically trained on. This highlights the additional benefit that can be ascertained from time-ordered corpora, with implications for temporal tasks in the era of widespread LLM use.
Reconstructing Sepsis Trajectories from Clinical Case Reports using LLMs: the Textual Time Series Corpus for Sepsis
Apr 12, 2025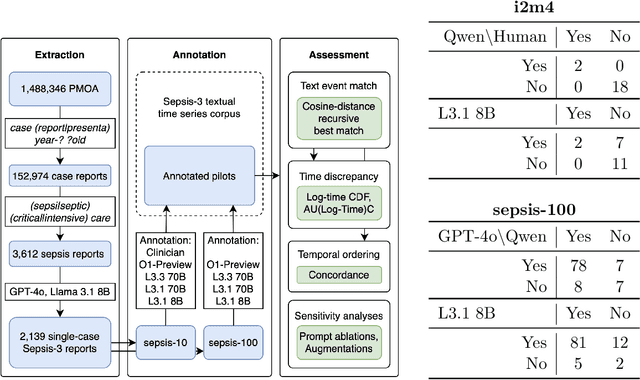
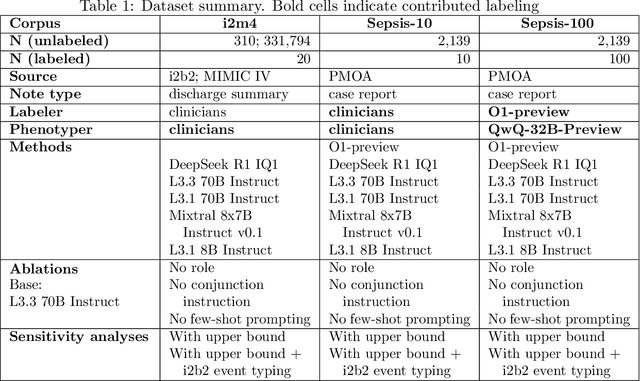
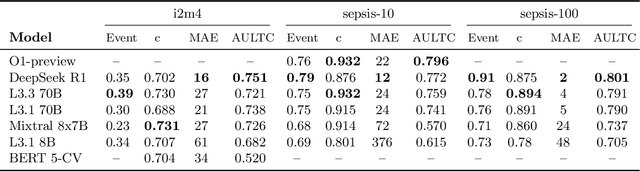
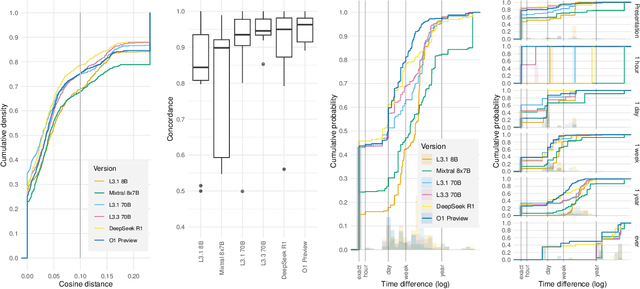
Abstract:Clinical case reports and discharge summaries may be the most complete and accurate summarization of patient encounters, yet they are finalized, i.e., timestamped after the encounter. Complementary data structured streams become available sooner but suffer from incompleteness. To train models and algorithms on more complete and temporally fine-grained data, we construct a pipeline to phenotype, extract, and annotate time-localized findings within case reports using large language models. We apply our pipeline to generate an open-access textual time series corpus for Sepsis-3 comprising 2,139 case reports from the Pubmed-Open Access (PMOA) Subset. To validate our system, we apply it on PMOA and timeline annotations from I2B2/MIMIC-IV and compare the results to physician-expert annotations. We show high recovery rates of clinical findings (event match rates: O1-preview--0.755, Llama 3.3 70B Instruct--0.753) and strong temporal ordering (concordance: O1-preview--0.932, Llama 3.3 70B Instruct--0.932). Our work characterizes the ability of LLMs to time-localize clinical findings in text, illustrating the limitations of LLM use for temporal reconstruction and providing several potential avenues of improvement via multimodal integration.
Temporal Supervised Contrastive Learning for Modeling Patient Risk Progression
Dec 10, 2023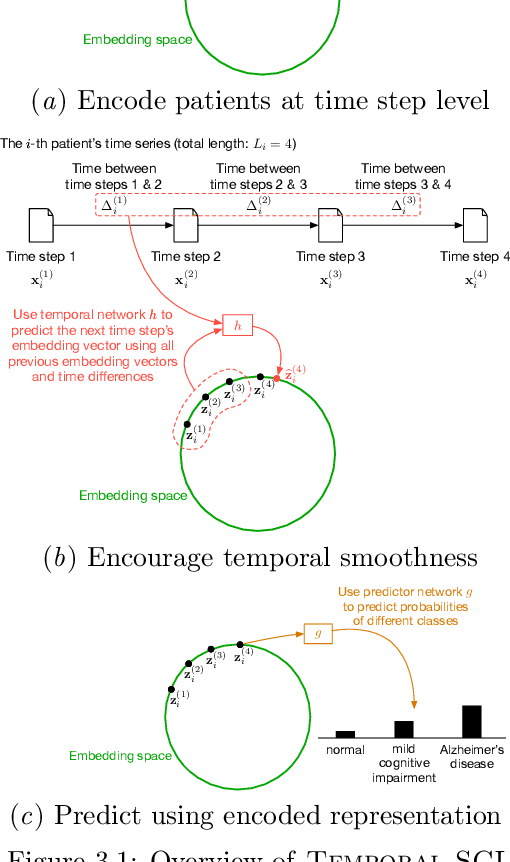
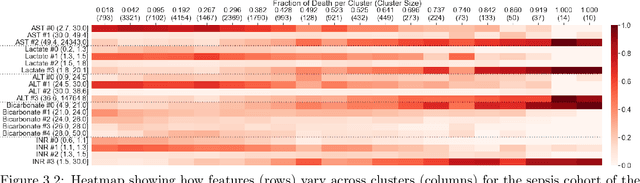
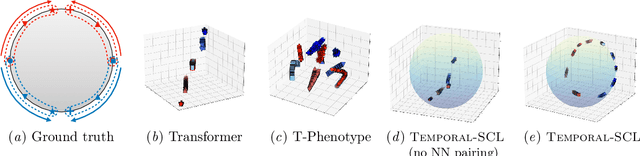
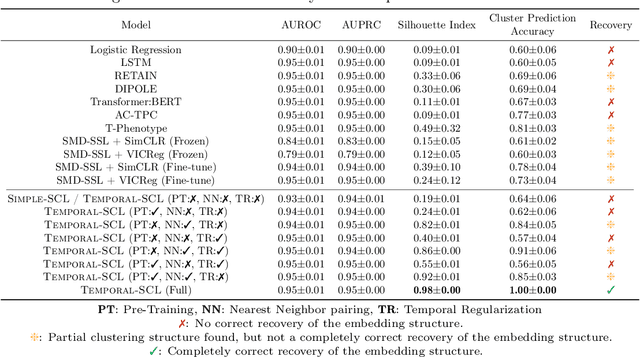
Abstract:We consider the problem of predicting how the likelihood of an outcome of interest for a patient changes over time as we observe more of the patient data. To solve this problem, we propose a supervised contrastive learning framework that learns an embedding representation for each time step of a patient time series. Our framework learns the embedding space to have the following properties: (1) nearby points in the embedding space have similar predicted class probabilities, (2) adjacent time steps of the same time series map to nearby points in the embedding space, and (3) time steps with very different raw feature vectors map to far apart regions of the embedding space. To achieve property (3), we employ a nearest neighbor pairing mechanism in the raw feature space. This mechanism also serves as an alternative to data augmentation, a key ingredient of contrastive learning, which lacks a standard procedure that is adequately realistic for clinical tabular data, to our knowledge. We demonstrate that our approach outperforms state-of-the-art baselines in predicting mortality of septic patients (MIMIC-III dataset) and tracking progression of cognitive impairment (ADNI dataset). Our method also consistently recovers the correct synthetic dataset embedding structure across experiments, a feat not achieved by baselines. Our ablation experiments show the pivotal role of our nearest neighbor pairing.
Fair Decision-making Under Uncertainty
Jan 29, 2023



Abstract:There has been concern within the artificial intelligence (AI) community and the broader society regarding the potential lack of fairness of AI-based decision-making systems. Surprisingly, there is little work quantifying and guaranteeing fairness in the presence of uncertainty which is prevalent in many socially sensitive applications, ranging from marketing analytics to actuarial analysis and recidivism prediction instruments. To this end, we study a longitudinal censored learning problem subject to fairness constraints, where we require that algorithmic decisions made do not affect certain individuals or social groups negatively in the presence of uncertainty on class label due to censorship. We argue that this formulation has a broader applicability to practical scenarios concerning fairness. We show how the newly devised fairness notions involving censored information and the general framework for fair predictions in the presence of censorship allow us to measure and mitigate discrimination under uncertainty that bridges the gap with real-world applications. Empirical evaluations on real-world discriminated datasets with censorship demonstrate the practicality of our approach.
Learning Clinical Concepts for Predicting Risk of Progression to Severe COVID-19
Aug 28, 2022



Abstract:With COVID-19 now pervasive, identification of high-risk individuals is crucial. Using data from a major healthcare provider in Southwestern Pennsylvania, we develop survival models predicting severe COVID-19 progression. In this endeavor, we face a tradeoff between more accurate models relying on many features and less accurate models relying on a few features aligned with clinician intuition. Complicating matters, many EHR features tend to be under-coded, degrading the accuracy of smaller models. In this study, we develop two sets of high-performance risk scores: (i) an unconstrained model built from all available features; and (ii) a pipeline that learns a small set of clinical concepts before training a risk predictor. Learned concepts boost performance over the corresponding features (C-index 0.858 vs. 0.844) and demonstrate improvements over (i) when evaluated out-of-sample (subsequent time periods). Our models outperform previous works (C-index 0.844-0.872 vs. 0.598-0.810).
Longitudinal Fairness with Censorship
Mar 31, 2022



Abstract:Recent works in artificial intelligence fairness attempt to mitigate discrimination by proposing constrained optimization programs that achieve parity for some fairness statistic. Most assume availability of the class label, which is impractical in many real-world applications such as precision medicine, actuarial analysis and recidivism prediction. Here we consider fairness in longitudinal right-censored environments, where the time to event might be unknown, resulting in censorship of the class label and inapplicability of existing fairness studies. We devise applicable fairness measures, propose a debiasing algorithm, and provide necessary theoretical constructs to bridge fairness with and without censorship for these important and socially-sensitive tasks. Our experiments on four censored datasets confirm the utility of our approach.
Fairness Amidst Non-IID Graph Data: A Literature Review
Feb 16, 2022

Abstract:Fairness in machine learning (ML), the process to understand and correct algorithmic bias, has gained increasing attention with numerous literature being carried out, commonly assume the underlying data is independent and identically distributed (IID). On the other hand, graphs are a ubiquitous data structure to capture connections among individual units and is non-IID by nature. It is therefore of great importance to bridge the traditional fairness literature designed on IID data and ubiquitous non-IID graph representations to tackle bias in ML systems. In this survey, we review such recent advance in fairness amidst non-IID graph data and identify datasets and evaluation metrics available for future research. We also point out the limitations of existing work as well as promising future directions.
FARF: A Fair and Adaptive Random Forests Classifier
Aug 21, 2021


Abstract:As Artificial Intelligence (AI) is used in more applications, the need to consider and mitigate biases from the learned models has followed. Most works in developing fair learning algorithms focus on the offline setting. However, in many real-world applications data comes in an online fashion and needs to be processed on the fly. Moreover, in practical application, there is a trade-off between accuracy and fairness that needs to be accounted for, but current methods often have multiple hyperparameters with non-trivial interaction to achieve fairness. In this paper, we propose a flexible ensemble algorithm for fair decision-making in the more challenging context of evolving online settings. This algorithm, called FARF (Fair and Adaptive Random Forests), is based on using online component classifiers and updating them according to the current distribution, that also accounts for fairness and a single hyperparameters that alters fairness-accuracy balance. Experiments on real-world discriminated data streams demonstrate the utility of FARF.
Neural Topic Models with Survival Supervision: Jointly Predicting Time-to-Event Outcomes and Learning How Clinical Features Relate
Jul 15, 2020
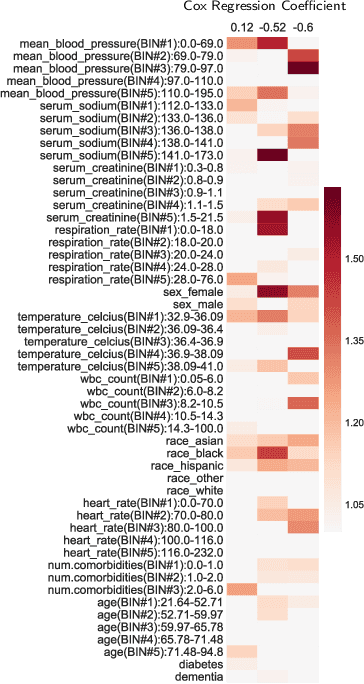
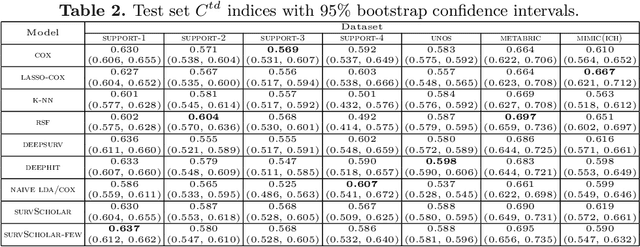
Abstract:In time-to-event prediction problems, a standard approach to estimating an interpretable model is to use Cox proportional hazards, where features are selected based on lasso regularization or stepwise regression. However, these Cox-based models do not learn how different features relate. As an alternative, we present an interpretable neural network approach to jointly learn a survival model to predict time-to-event outcomes while simultaneously learning how features relate in terms of a topic model. In particular, we model each subject as a distribution over "topics", which are learned from clinical features as to help predict a time-to-event outcome. From a technical standpoint, we extend existing neural topic modeling approaches to also minimize a survival analysis loss function. We study the effectiveness of this approach on seven healthcare datasets on predicting time until death as well as hospital ICU length of stay, where we find that neural survival-supervised topic models achieves competitive accuracy with existing approaches while yielding interpretable clinical "topics" that explain feature relationships.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge