Shahriar Noroozizadeh
Deep sequence models tend to memorize geometrically; it is unclear why
Oct 30, 2025Abstract:In sequence modeling, the parametric memory of atomic facts has been predominantly abstracted as a brute-force lookup of co-occurrences between entities. We contrast this associative view against a geometric view of how memory is stored. We begin by isolating a clean and analyzable instance of Transformer reasoning that is incompatible with memory as strictly a storage of the local co-occurrences specified during training. Instead, the model must have somehow synthesized its own geometry of atomic facts, encoding global relationships between all entities, including non-co-occurring ones. This in turn has simplified a hard reasoning task involving an $\ell$-fold composition into an easy-to-learn 1-step geometric task. From this phenomenon, we extract fundamental aspects of neural embedding geometries that are hard to explain. We argue that the rise of such a geometry, despite optimizing over mere local associations, cannot be straightforwardly attributed to typical architectural or optimizational pressures. Counterintuitively, an elegant geometry is learned even when it is not more succinct than a brute-force lookup of associations. Then, by analyzing a connection to Node2Vec, we demonstrate how the geometry stems from a spectral bias that -- in contrast to prevailing theories -- indeed arises naturally despite the lack of various pressures. This analysis also points to practitioners a visible headroom to make Transformer memory more strongly geometric. We hope the geometric view of parametric memory encourages revisiting the default intuitions that guide researchers in areas like knowledge acquisition, capacity, discovery and unlearning.
Forecasting from Clinical Textual Time Series: Adaptations of the Encoder and Decoder Language Model Families
Apr 14, 2025Abstract:Clinical case reports encode rich, temporal patient trajectories that are often underexploited by traditional machine learning methods relying on structured data. In this work, we introduce the forecasting problem from textual time series, where timestamped clinical findings--extracted via an LLM-assisted annotation pipeline--serve as the primary input for prediction. We systematically evaluate a diverse suite of models, including fine-tuned decoder-based large language models and encoder-based transformers, on tasks of event occurrence prediction, temporal ordering, and survival analysis. Our experiments reveal that encoder-based models consistently achieve higher F1 scores and superior temporal concordance for short- and long-horizon event forecasting, while fine-tuned masking approaches enhance ranking performance. In contrast, instruction-tuned decoder models demonstrate a relative advantage in survival analysis, especially in early prognosis settings. Our sensitivity analyses further demonstrate the importance of time ordering, which requires clinical time series construction, as compared to text ordering, the format of the text inputs that LLMs are classically trained on. This highlights the additional benefit that can be ascertained from time-ordered corpora, with implications for temporal tasks in the era of widespread LLM use.
Reconstructing Sepsis Trajectories from Clinical Case Reports using LLMs: the Textual Time Series Corpus for Sepsis
Apr 12, 2025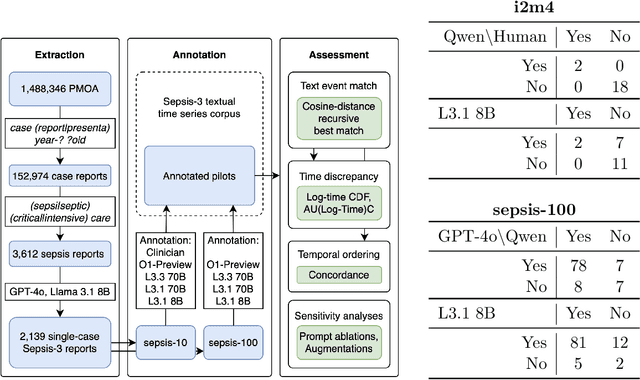
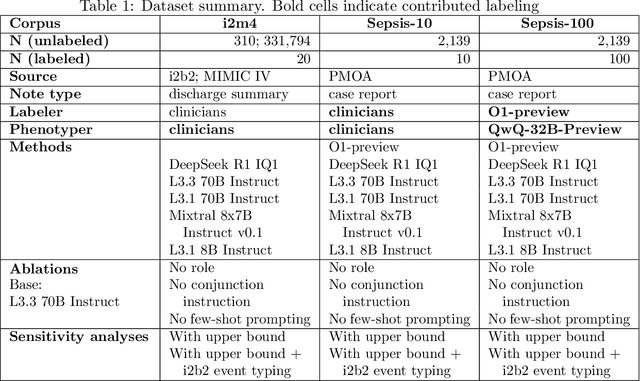
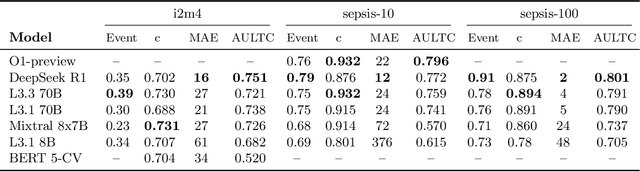
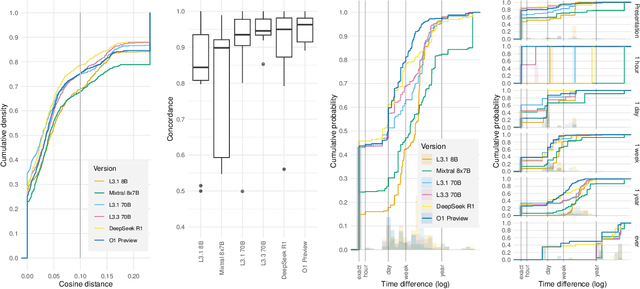
Abstract:Clinical case reports and discharge summaries may be the most complete and accurate summarization of patient encounters, yet they are finalized, i.e., timestamped after the encounter. Complementary data structured streams become available sooner but suffer from incompleteness. To train models and algorithms on more complete and temporally fine-grained data, we construct a pipeline to phenotype, extract, and annotate time-localized findings within case reports using large language models. We apply our pipeline to generate an open-access textual time series corpus for Sepsis-3 comprising 2,139 case reports from the Pubmed-Open Access (PMOA) Subset. To validate our system, we apply it on PMOA and timeline annotations from I2B2/MIMIC-IV and compare the results to physician-expert annotations. We show high recovery rates of clinical findings (event match rates: O1-preview--0.755, Llama 3.3 70B Instruct--0.753) and strong temporal ordering (concordance: O1-preview--0.932, Llama 3.3 70B Instruct--0.932). Our work characterizes the ability of LLMs to time-localize clinical findings in text, illustrating the limitations of LLM use for temporal reconstruction and providing several potential avenues of improvement via multimodal integration.
Many-Shot In-Context Learning for Molecular Inverse Design
Jul 26, 2024Abstract:Large Language Models (LLMs) have demonstrated great performance in few-shot In-Context Learning (ICL) for a variety of generative and discriminative chemical design tasks. The newly expanded context windows of LLMs can further improve ICL capabilities for molecular inverse design and lead optimization. To take full advantage of these capabilities we developed a new semi-supervised learning method that overcomes the lack of experimental data available for many-shot ICL. Our approach involves iterative inclusion of LLM generated molecules with high predicted performance, along with experimental data. We further integrated our method in a multi-modal LLM which allows for the interactive modification of generated molecular structures using text instructions. As we show, the new method greatly improves upon existing ICL methods for molecular design while being accessible and easy to use for scientists.
ET tu, CLIP? Addressing Common Object Errors for Unseen Environments
Jun 25, 2024Abstract:We introduce a simple method that employs pre-trained CLIP encoders to enhance model generalization in the ALFRED task. In contrast to previous literature where CLIP replaces the visual encoder, we suggest using CLIP as an additional module through an auxiliary object detection objective. We validate our method on the recently proposed Episodic Transformer architecture and demonstrate that incorporating CLIP improves task performance on the unseen validation set. Additionally, our analysis results support that CLIP especially helps with leveraging object descriptions, detecting small objects, and interpreting rare words.
TLDR at SemEval-2024 Task 2: T5-generated clinical-Language summaries for DeBERTa Report Analysis
Apr 14, 2024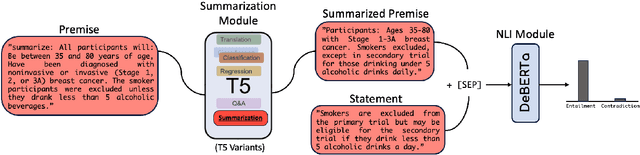
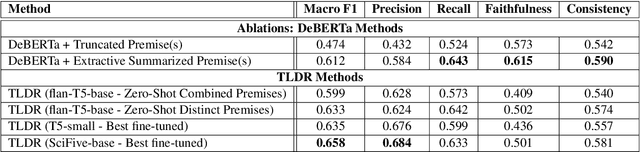
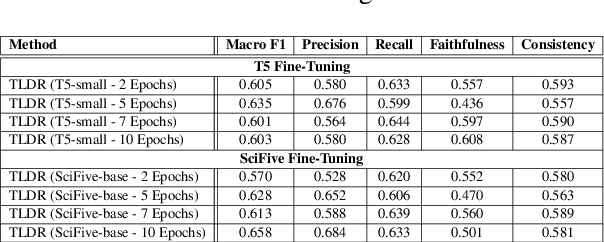
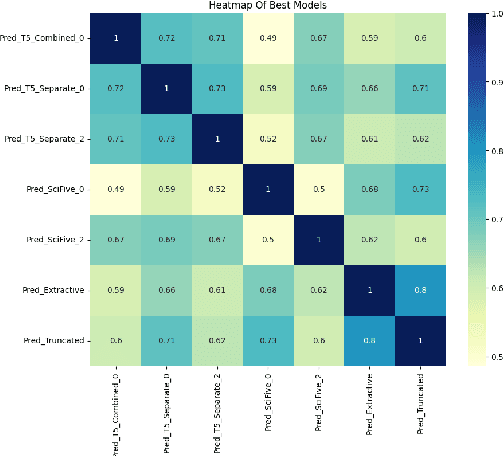
Abstract:This paper introduces novel methodologies for the Natural Language Inference for Clinical Trials (NLI4CT) task. We present TLDR (T5-generated clinical-Language summaries for DeBERTa Report Analysis) which incorporates T5-model generated premise summaries for improved entailment and contradiction analysis in clinical NLI tasks. This approach overcomes the challenges posed by small context windows and lengthy premises, leading to a substantial improvement in Macro F1 scores: a 0.184 increase over truncated premises. Our comprehensive experimental evaluation, including detailed error analysis and ablations, confirms the superiority of TLDR in achieving consistency and faithfulness in predictions against semantically altered inputs.
Temporal Supervised Contrastive Learning for Modeling Patient Risk Progression
Dec 10, 2023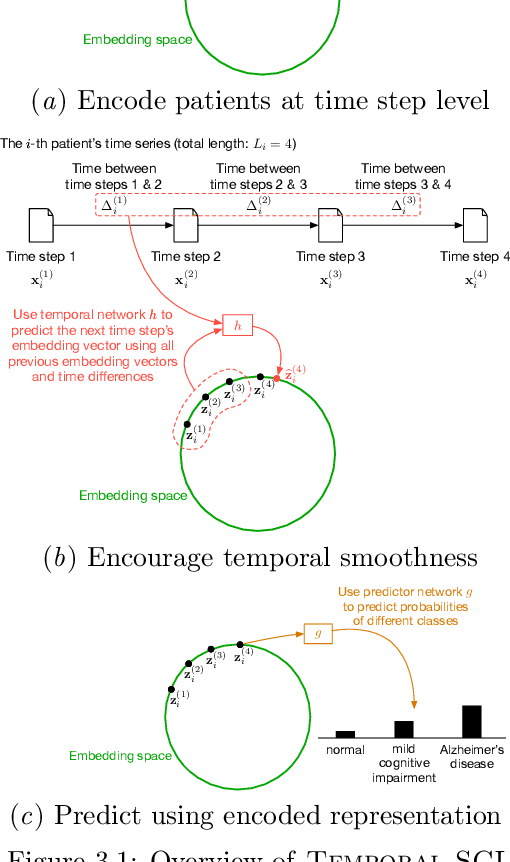
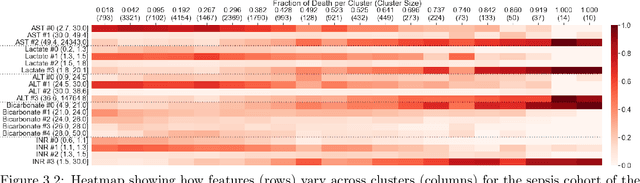
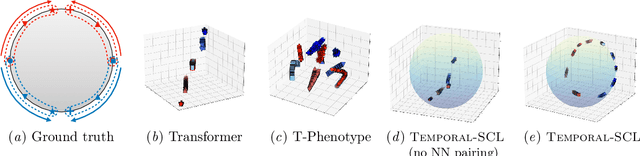
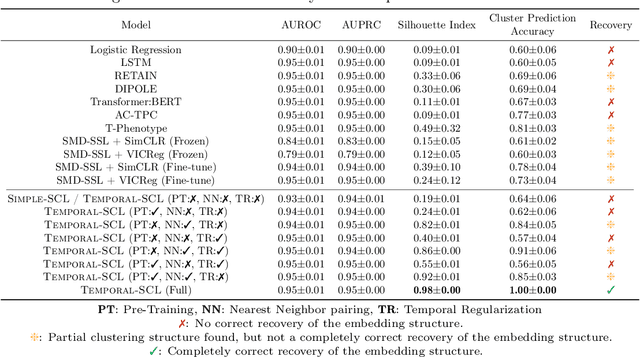
Abstract:We consider the problem of predicting how the likelihood of an outcome of interest for a patient changes over time as we observe more of the patient data. To solve this problem, we propose a supervised contrastive learning framework that learns an embedding representation for each time step of a patient time series. Our framework learns the embedding space to have the following properties: (1) nearby points in the embedding space have similar predicted class probabilities, (2) adjacent time steps of the same time series map to nearby points in the embedding space, and (3) time steps with very different raw feature vectors map to far apart regions of the embedding space. To achieve property (3), we employ a nearest neighbor pairing mechanism in the raw feature space. This mechanism also serves as an alternative to data augmentation, a key ingredient of contrastive learning, which lacks a standard procedure that is adequately realistic for clinical tabular data, to our knowledge. We demonstrate that our approach outperforms state-of-the-art baselines in predicting mortality of septic patients (MIMIC-III dataset) and tracking progression of cognitive impairment (ADNI dataset). Our method also consistently recovers the correct synthetic dataset embedding structure across experiments, a feat not achieved by baselines. Our ablation experiments show the pivotal role of our nearest neighbor pairing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge