Gang Liu
Enhancing Automatic Chord Recognition via Pseudo-Labeling and Knowledge Distillation
Feb 23, 2026Abstract:Automatic Chord Recognition (ACR) is constrained by the scarcity of aligned chord labels, as well-aligned annotations are costly to acquire. At the same time, open-weight pre-trained models are currently more accessible than their proprietary training data. In this work, we present a two-stage training pipeline that leverages pre-trained models together with unlabeled audio. The proposed method decouples training into two stages. In the first stage, we use a pre-trained BTC model as a teacher to generate pseudo-labels for over 1,000 hours of diverse unlabeled audio and train a student model solely on these pseudo-labels. In the second stage, the student is continually trained on ground-truth labels as they become available, with selective knowledge distillation (KD) from the teacher applied as a regularizer to prevent catastrophic forgetting of the representations learned in the first stage. In our experiments, two models (BTC, 2E1D) were used as students. In stage 1, using only pseudo-labels, the BTC student achieves over 98% of the teacher's performance, while the 2E1D model achieves about 96% across seven standard mir_eval metrics. After a single training run for both students in stage 2, the resulting BTC student model surpasses the traditional supervised learning baseline by 2.5% and the original pre-trained teacher model by 1.55% on average across all metrics. And the resulting 2E1D student model improves from the traditional supervised learning baseline by 3.79% on average and achieves almost the same performance as the teacher. Both cases show the large gains on rare chord qualities.
Adaptive Global and Fine-Grained Perceptual Fusion for MLLM Embeddings Compatible with Hard Negative Amplification
Feb 05, 2026Abstract:Multimodal embeddings serve as a bridge for aligning vision and language, with the two primary implementations -- CLIP-based and MLLM-based embedding models -- both limited to capturing only global semantic information. Although numerous studies have focused on fine-grained understanding, we observe that complex scenarios currently targeted by MLLM embeddings often involve a hybrid perceptual pattern of both global and fine-grained elements, thus necessitating a compatible fusion mechanism. In this paper, we propose Adaptive Global and Fine-grained perceptual Fusion for MLLM Embeddings (AGFF-Embed), a method that prompts the MLLM to generate multiple embeddings focusing on different dimensions of semantic information, which are then adaptively and smoothly aggregated. Furthermore, we adapt AGFF-Embed with the Explicit Gradient Amplification (EGA) technique to achieve in-batch hard negatives enhancement without requiring fine-grained editing of the dataset. Evaluation on the MMEB and MMVP-VLM benchmarks shows that AGFF-Embed comprehensively achieves state-of-the-art performance in both general and fine-grained understanding compared to other multimodal embedding models.
IVC-Prune: Revealing the Implicit Visual Coordinates in LVLMs for Vision Token Pruning
Feb 03, 2026Abstract:Large Vision-Language Models (LVLMs) achieve impressive performance across multiple tasks. A significant challenge, however, is their prohibitive inference cost when processing high-resolution visual inputs. While visual token pruning has emerged as a promising solution, existing methods that primarily focus on semantic relevance often discard tokens that are crucial for spatial reasoning. We address this gap through a novel insight into \emph{how LVLMs process spatial reasoning}. Specifically, we reveal that LVLMs implicitly establish visual coordinate systems through Rotary Position Embeddings (RoPE), where specific token positions serve as \textbf{implicit visual coordinates} (IVC tokens) that are essential for spatial reasoning. Based on this insight, we propose \textbf{IVC-Prune}, a training-free, prompt-aware pruning strategy that retains both IVC tokens and semantically relevant foreground tokens. IVC tokens are identified by theoretically analyzing the mathematical properties of RoPE, targeting positions at which its rotation matrices approximate identity matrix or the $90^\circ$ rotation matrix. Foreground tokens are identified through a robust two-stage process: semantic seed discovery followed by contextual refinement via value-vector similarity. Extensive evaluations across four representative LVLMs and twenty diverse benchmarks show that IVC-Prune reduces visual tokens by approximately 50\% while maintaining $\geq$ 99\% of the original performance and even achieving improvements on several benchmarks. Source codes are available at https://github.com/FireRedTeam/IVC-Prune.
AutoHealth: An Uncertainty-Aware Multi-Agent System for Autonomous Health Data Modeling
Feb 01, 2026Abstract:LLM-based agents have demonstrated strong potential for autonomous machine learning, yet their applicability to health data remains limited. Existing systems often struggle to generalize across heterogeneous health data modalities, rely heavily on predefined solution templates with insufficient adaptation to task-specific objectives, and largely overlook uncertainty estimation, which is essential for reliable decision-making in healthcare. To address these challenges, we propose \textit{AutoHealth}, a novel uncertainty-aware multi-agent system that autonomously models health data and assesses model reliability. \textit{AutoHealth} employs closed-loop coordination among five specialized agents to perform data exploration, task-conditioned model construction, training, and optimization, while jointly prioritizing predictive performance and uncertainty quantification. Beyond producing ready-to-use models, the system generates comprehensive reports to support trustworthy interpretation and risk-aware decision-making. To rigorously evaluate its effectiveness, we curate a challenging real-world benchmark comprising 17 tasks across diverse data modalities and learning settings. \textit{AutoHealth} completes all tasks and outperforms state-of-the-art baselines by 29.2\% in prediction performance and 50.2\% in uncertainty estimation.
Concise Geometric Description as a Bridge: Unleashing the Potential of LLM for Plane Geometry Problem Solving
Jan 29, 2026Abstract:Plane Geometry Problem Solving (PGPS) is a multimodal reasoning task that aims to solve a plane geometric problem based on a geometric diagram and problem textual descriptions. Although Large Language Models (LLMs) possess strong reasoning skills, their direct application to PGPS is hindered by their inability to process visual diagrams. Existing works typically fine-tune Multimodal LLMs (MLLMs) end-to-end on large-scale PGPS data to enhance visual understanding and reasoning simultaneously. However, such joint optimization may compromise base LLMs' inherent reasoning capability. In this work, we observe that LLM itself is potentially a powerful PGPS solver when appropriately formulating visual information as textual descriptions. We propose to train a MLLM Interpreter to generate geometric descriptions for the visual diagram, and an off-the-shelf LLM is utilized to perform reasoning. Specifically, we choose Conditional Declaration Language (CDL) as the geometric description as its conciseness eases the MLLM Interpreter training. The MLLM Interpreter is fine-tuned via CoT (Chain-of-Thought)-augmented SFT followed by GRPO to generate CDL. Instead of using a conventional solution-based reward that compares the reasoning result with the ground-truth answer, we design CDL matching rewards to facilitate more effective GRPO training, which provides more direct and denser guidance for CDL generation. To support training, we construct a new dataset, Formalgeo7k-Rec-CoT, by manually reviewing Formalgeo7k v2 and incorporating CoT annotations. Extensive experiments on Formalgeo7k-Rec-CoT, Unigeo, and MathVista show our method (finetuned on only 5.5k data) performs favorably against leading open-source and closed-source MLLMs.
Next Generation Active Learning: Mixture of LLMs in the Loop
Jan 22, 2026Abstract:With the rapid advancement and strong generalization capabilities of large language models (LLMs), they have been increasingly incorporated into the active learning pipelines as annotators to reduce annotation costs. However, considering the annotation quality, labels generated by LLMs often fall short of real-world applicability. To address this, we propose a novel active learning framework, Mixture of LLMs in the Loop Active Learning, replacing human annotators with labels generated through a Mixture-of-LLMs-based annotation model, aimed at enhancing LLM-based annotation robustness by aggregating the strengths of multiple LLMs. To further mitigate the impact of the noisy labels, we introduce annotation discrepancy and negative learning to identify the unreliable annotations and enhance learning effectiveness. Extensive experiments demonstrate that our framework achieves performance comparable to human annotation and consistently outperforms single-LLM baselines and other LLM-ensemble-based approaches. Moreover, our framework is built on lightweight LLMs, enabling it to operate fully on local machines in real-world applications.
Sequence of Expert: Boosting Imitation Planners for Autonomous Driving through Temporal Alternation
Dec 15, 2025Abstract:Imitation learning (IL) has emerged as a central paradigm in autonomous driving. While IL excels in matching expert behavior in open-loop settings by minimizing per-step prediction errors, its performance degrades unexpectedly in closed-loop due to the gradual accumulation of small, often imperceptible errors over time.Over successive planning cycles, these errors compound, potentially resulting in severe failures.Current research efforts predominantly rely on increasingly sophisticated network architectures or high-fidelity training datasets to enhance the robustness of IL planners against error accumulation, focusing on the state-level robustness at a single time point. However, autonomous driving is inherently a continuous-time process, and leveraging the temporal scale to enhance robustness may provide a new perspective for addressing this issue.To this end, we propose a method termed Sequence of Experts (SoE), a temporal alternation policy that enhances closed-loop performance without increasing model size or data requirements. Our experiments on large-scale autonomous driving benchmarks nuPlan demonstrate that SoE method consistently and significantly improves the performance of all the evaluated models, and achieves state-of-the-art performance.This module may provide a key and widely applicable support for improving the training efficiency of autonomous driving models.
Open Polymer Challenge: Post-Competition Report
Dec 09, 2025Abstract:Machine learning (ML) offers a powerful path toward discovering sustainable polymer materials, but progress has been limited by the lack of large, high-quality, and openly accessible polymer datasets. The Open Polymer Challenge (OPC) addresses this gap by releasing the first community-developed benchmark for polymer informatics, featuring a dataset with 10K polymers and 5 properties: thermal conductivity, radius of gyration, density, fractional free volume, and glass transition temperature. The challenge centers on multi-task polymer property prediction, a core step in virtual screening pipelines for materials discovery. Participants developed models under realistic constraints that include small data, label imbalance, and heterogeneous simulation sources, using techniques such as feature-based augmentation, transfer learning, self-supervised pretraining, and targeted ensemble strategies. The competition also revealed important lessons about data preparation, distribution shifts, and cross-group simulation consistency, informing best practices for future large-scale polymer datasets. The resulting models, analysis, and released data create a new foundation for molecular AI in polymer science and are expected to accelerate the development of sustainable and energy-efficient materials. Along with the competition, we release the test dataset at https://www.kaggle.com/datasets/alexliu99/neurips-open-polymer-prediction-2025-test-data. We also release the data generation pipeline at https://github.com/sobinalosious/ADEPT, which simulates more than 25 properties, including thermal conductivity, radius of gyration, and density.
Protein Structure Tokenization via Geometric Byte Pair Encoding
Nov 13, 2025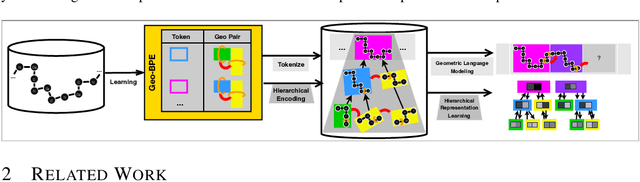
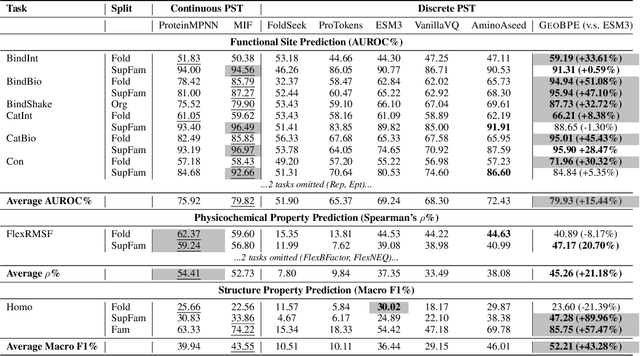
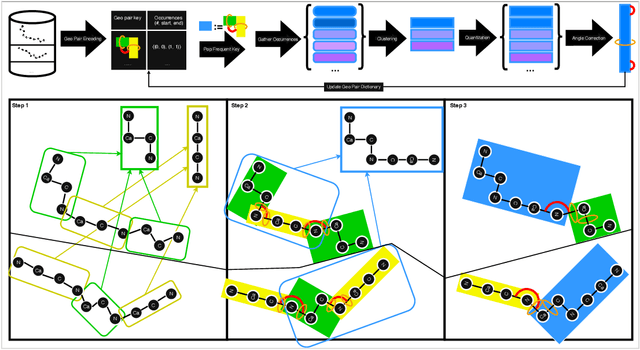
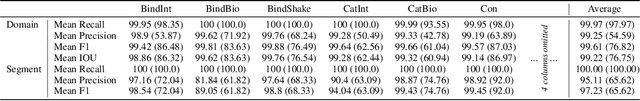
Abstract:Protein structure is central to biological function, and enabling multimodal protein models requires joint reasoning over sequence, structure, and function. A key barrier is the lack of principled protein structure tokenizers (PSTs): existing approaches fix token size or rely on continuous vector codebooks, limiting interpretability, multi-scale control, and transfer across architectures. We introduce GeoBPE, a geometry-grounded PST that transforms continuous, noisy, multi-scale backbone conformations into discrete ``sentences'' of geometry while enforcing global constraints. Analogous to byte-pair encoding, GeoBPE generates a hierarchical vocabulary of geometric primitives by iteratively (i) clustering Geo-Pair occurrences with k-medoids to yield a resolution-controllable vocabulary; (ii) quantizing each Geo-Pair to its closest medoid prototype; and (iii) reducing drift through differentiable inverse kinematics that optimizes boundary glue angles under an $\mathrm{SE}(3)$ end-frame loss. GeoBPE offers compression ($>$10x reduction in bits-per-residue at similar distortion rate), data efficiency ($>$10x less training data), and generalization (maintains test/train distortion ratio of $1.0-1.1$). It is architecture-agnostic: (a) its hierarchical vocabulary provides a strong inductive bias for coarsening residue-level embeddings from large PLMs into motif- and protein-level representations, consistently outperforming leading PSTs across $12$ tasks and $24$ test splits; (b) paired with a transformer, GeoBPE supports unconditional backbone generation via language modeling; and (c) tokens align with CATH functional families and support expert-interpretable case studies, offering functional meaning absent in prior PSTs. Code is available at https://github.com/shiningsunnyday/PT-BPE/.
Learning to Pose Problems: Reasoning-Driven and Solver-Adaptive Data Synthesis for Large Reasoning Models
Nov 13, 2025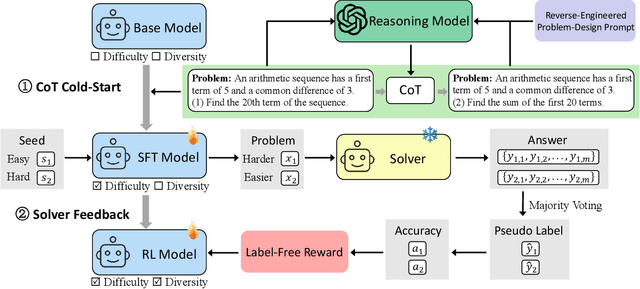
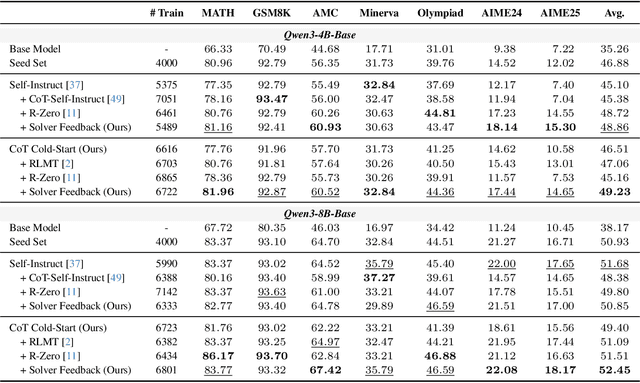

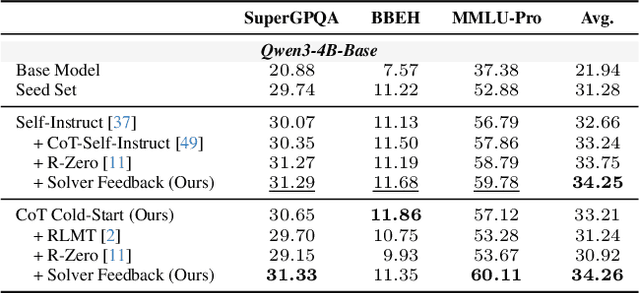
Abstract:Data synthesis for training large reasoning models offers a scalable alternative to limited, human-curated datasets, enabling the creation of high-quality data. However, existing approaches face several challenges: (i) indiscriminate generation that ignores the solver's ability and yields low-value problems, or reliance on complex data pipelines to balance problem difficulty; and (ii) a lack of reasoning in problem generation, leading to shallow problem variants. In this paper, we develop a problem generator that reasons explicitly to plan problem directions before synthesis and adapts difficulty to the solver's ability. Specifically, we construct related problem pairs and augment them with intermediate problem-design CoT produced by a reasoning model. These data bootstrap problem-design strategies from the generator. Then, we treat the solver's feedback on synthetic problems as a reward signal, enabling the generator to calibrate difficulty and produce complementary problems near the edge of the solver's competence. Extensive experiments on 10 mathematical and general reasoning benchmarks show that our method achieves an average improvement of 2.5% and generalizes to both language and vision-language models. Moreover, a solver trained on the synthesized data provides improved rewards for continued generator training, enabling co-evolution and yielding a further 0.7% performance gain. Our code will be made publicly available here.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge