David Buckeridge
MixEHR-Nest: Identifying Subphenotypes within Electronic Health Records through Hierarchical Guided-Topic Modeling
Oct 17, 2024



Abstract:Automatic subphenotyping from electronic health records (EHRs)provides numerous opportunities to understand diseases with unique subgroups and enhance personalized medicine for patients. However, existing machine learning algorithms either focus on specific diseases for better interpretability or produce coarse-grained phenotype topics without considering nuanced disease patterns. In this study, we propose a guided topic model, MixEHR-Nest, to infer sub-phenotype topics from thousands of disease using multi-modal EHR data. Specifically, MixEHR-Nest detects multiple subtopics from each phenotype topic, whose prior is guided by the expert-curated phenotype concepts such as Phenotype Codes (PheCodes) or Clinical Classification Software (CCS) codes. We evaluated MixEHR-Nest on two EHR datasets: (1) the MIMIC-III dataset consisting of over 38 thousand patients from intensive care unit (ICU) from Beth Israel Deaconess Medical Center (BIDMC) in Boston, USA; (2) the healthcare administrative database PopHR, comprising 1.3 million patients from Montreal, Canada. Experimental results demonstrate that MixEHR-Nest can identify subphenotypes with distinct patterns within each phenotype, which are predictive for disease progression and severity. Consequently, MixEHR-Nest distinguishes between type 1 and type 2 diabetes by inferring subphenotypes using CCS codes, which do not differentiate these two subtype concepts. Additionally, MixEHR-Nest not only improved the prediction accuracy of short-term mortality of ICU patients and initial insulin treatment in diabetic patients but also revealed the contributions of subphenotypes. For longitudinal analysis, MixEHR-Nest identified subphenotypes of distinct age prevalence under the same phenotypes, such as asthma, leukemia, epilepsy, and depression. The MixEHR-Nest software is available at GitHub: https://github.com/li-lab-mcgill/MixEHR-Nest.
TrajGPT: Irregular Time-Series Representation Learning for Health Trajectory Analysis
Oct 03, 2024



Abstract:In many domains, such as healthcare, time-series data is often irregularly sampled with varying intervals between observations. This poses challenges for classical time-series models that require equally spaced data. To address this, we propose a novel time-series Transformer called Trajectory Generative Pre-trained Transformer (TrajGPT). TrajGPT employs a novel Selective Recurrent Attention (SRA) mechanism, which utilizes a data-dependent decay to adaptively filter out irrelevant past information based on contexts. By interpreting TrajGPT as discretized ordinary differential equations (ODEs), it effectively captures the underlying continuous dynamics and enables time-specific inference for forecasting arbitrary target timesteps. Experimental results demonstrate that TrajGPT excels in trajectory forecasting, drug usage prediction, and phenotype classification without requiring task-specific fine-tuning. By evolving the learned continuous dynamics, TrajGPT can interpolate and extrapolate disease risk trajectories from partially-observed time series. The visualization of predicted health trajectories shows that TrajGPT forecasts unseen diseases based on the history of clinically relevant phenotypes (i.e., contexts).
BAND: Biomedical Alert News Dataset
May 23, 2023
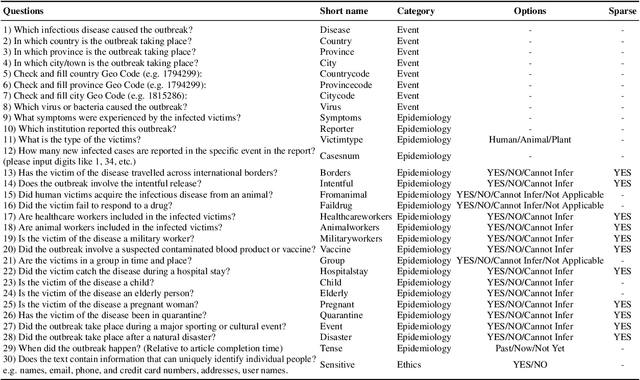
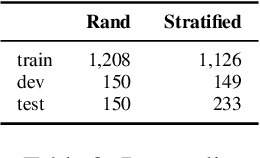
Abstract:Infectious disease outbreaks continue to pose a significant threat to human health and well-being. To improve disease surveillance and understanding of disease spread, several surveillance systems have been developed to monitor daily news alerts and social media. However, existing systems lack thorough epidemiological analysis in relation to corresponding alerts or news, largely due to the scarcity of well-annotated reports data. To address this gap, we introduce the Biomedical Alert News Dataset (BAND), which includes 1,508 samples from existing reported news articles, open emails, and alerts, as well as 30 epidemiology-related questions. These questions necessitate the model's expert reasoning abilities, thereby offering valuable insights into the outbreak of the disease. The BAND dataset brings new challenges to the NLP world, requiring better disguise capability of the content and the ability to infer important information. We provide several benchmark tasks, including Named Entity Recognition (NER), Question Answering (QA), and Event Extraction (EE), to show how existing models are capable of handling these tasks in the epidemiology domain. To the best of our knowledge, the BAND corpus is the largest corpus of well-annotated biomedical outbreak alert news with elaborately designed questions, making it a valuable resource for epidemiologists and NLP researchers alike.
Modeling electronic health record data using a knowledge-graph-embedded topic model
Jun 03, 2022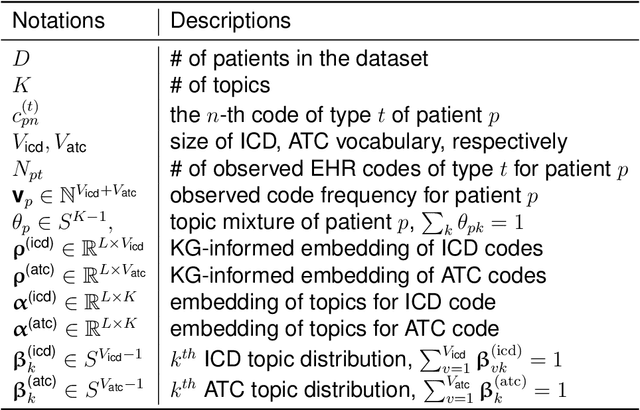
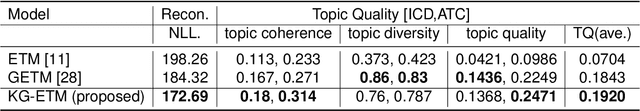
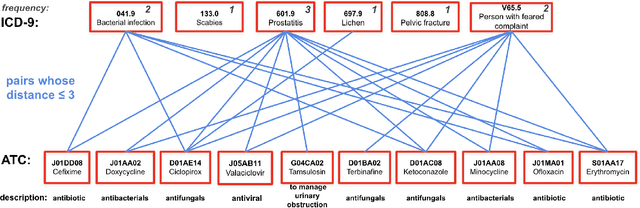
Abstract:The rapid growth of electronic health record (EHR) datasets opens up promising opportunities to understand human diseases in a systematic way. However, effective extraction of clinical knowledge from the EHR data has been hindered by its sparsity and noisy information. We present KG-ETM, an end-to-end knowledge graph-based multimodal embedded topic model. KG-ETM distills latent disease topics from EHR data by learning the embedding from the medical knowledge graphs. We applied KG-ETM to a large-scale EHR dataset consisting of over 1 million patients. We evaluated its performance based on EHR reconstruction and drug imputation. KG-ETM demonstrated superior performance over the alternative methods on both tasks. Moreover, our model learned clinically meaningful graph-informed embedding of the EHR codes. In additional, our model is also able to discover interpretable and accurate patient representations for patient stratification and drug recommendations.
Supervised multi-specialist topic model with applications on large-scale electronic health record data
May 04, 2021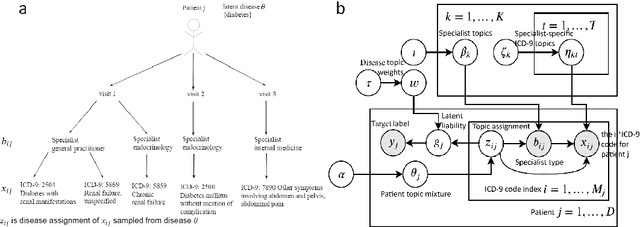
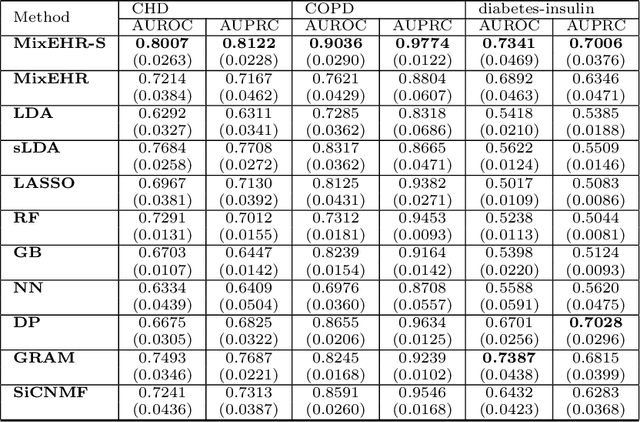
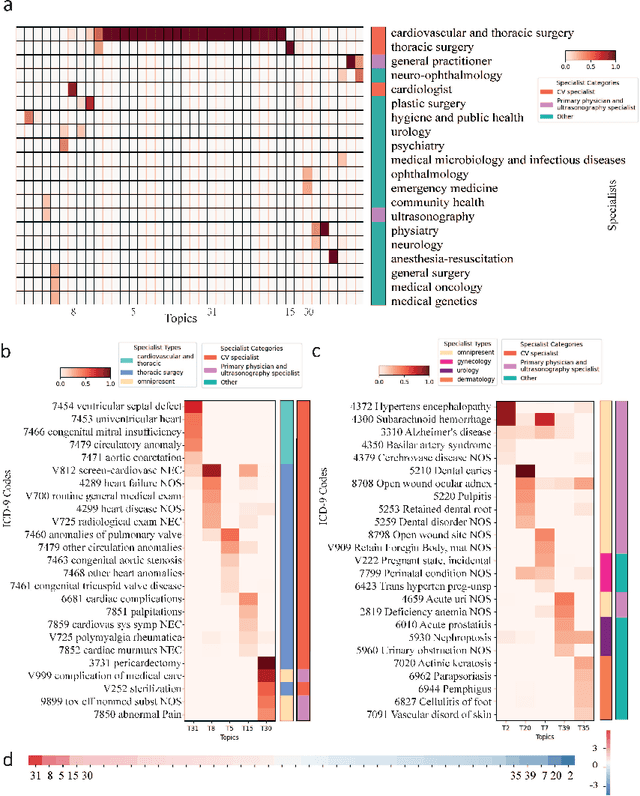
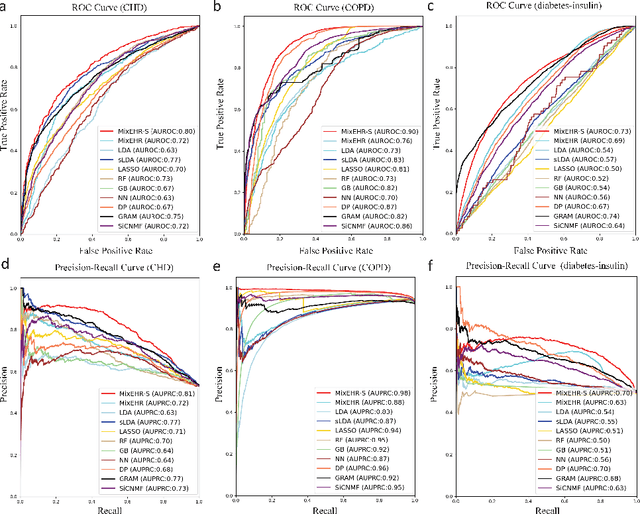
Abstract:Motivation: Electronic health record (EHR) data provides a new venue to elucidate disease comorbidities and latent phenotypes for precision medicine. To fully exploit its potential, a realistic data generative process of the EHR data needs to be modelled. We present MixEHR-S to jointly infer specialist-disease topics from the EHR data. As the key contribution, we model the specialist assignments and ICD-coded diagnoses as the latent topics based on patient's underlying disease topic mixture in a novel unified supervised hierarchical Bayesian topic model. For efficient inference, we developed a closed-form collapsed variational inference algorithm to learn the model distributions of MixEHR-S. We applied MixEHR-S to two independent large-scale EHR databases in Quebec with three targeted applications: (1) Congenital Heart Disease (CHD) diagnostic prediction among 154,775 patients; (2) Chronic obstructive pulmonary disease (COPD) diagnostic prediction among 73,791 patients; (3) future insulin treatment prediction among 78,712 patients diagnosed with diabetes as a mean to assess the disease exacerbation. In all three applications, MixEHR-S conferred clinically meaningful latent topics among the most predictive latent topics and achieved superior target prediction accuracy compared to the existing methods, providing opportunities for prioritizing high-risk patients for healthcare services. MixEHR-S source code and scripts of the experiments are freely available at https://github.com/li-lab-mcgill/mixehrS
COVI-AgentSim: an Agent-based Model for Evaluating Methods of Digital Contact Tracing
Oct 30, 2020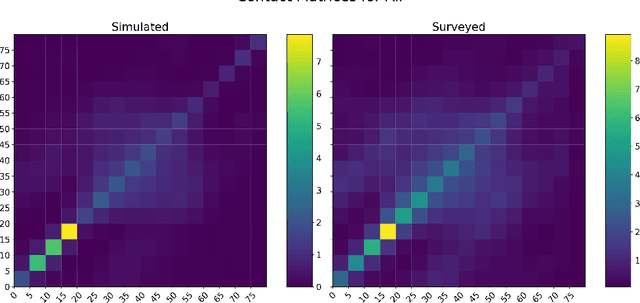
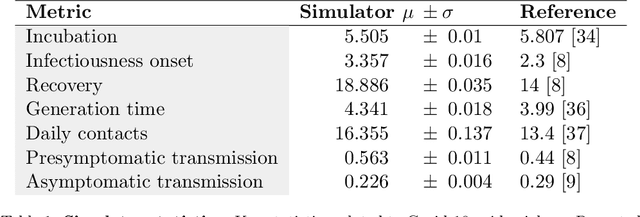
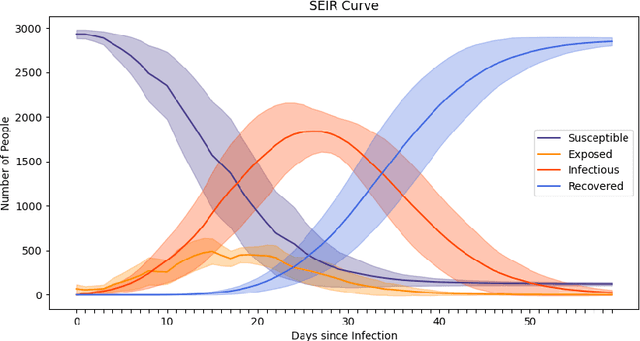
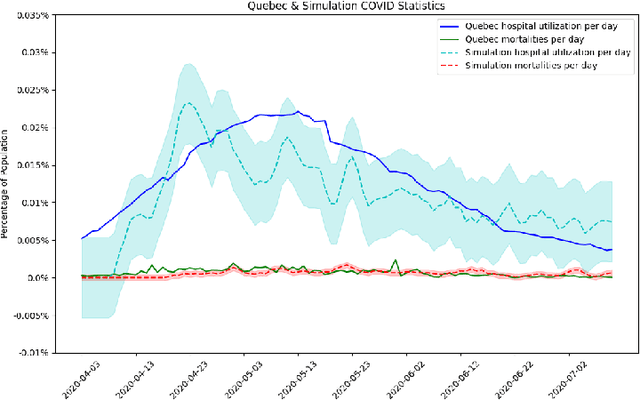
Abstract:The rapid global spread of COVID-19 has led to an unprecedented demand for effective methods to mitigate the spread of the disease, and various digital contact tracing (DCT) methods have emerged as a component of the solution. In order to make informed public health choices, there is a need for tools which allow evaluation and comparison of DCT methods. We introduce an agent-based compartmental simulator we call COVI-AgentSim, integrating detailed consideration of virology, disease progression, social contact networks, and mobility patterns, based on parameters derived from empirical research. We verify by comparing to real data that COVI-AgentSim is able to reproduce realistic COVID-19 spread dynamics, and perform a sensitivity analysis to verify that the relative performance of contact tracing methods are consistent across a range of settings. We use COVI-AgentSim to perform cost-benefit analyses comparing no DCT to: 1) standard binary contact tracing (BCT) that assigns binary recommendations based on binary test results; and 2) a rule-based method for feature-based contact tracing (FCT) that assigns a graded level of recommendation based on diverse individual features. We find all DCT methods consistently reduce the spread of the disease, and that the advantage of FCT over BCT is maintained over a wide range of adoption rates. Feature-based methods of contact tracing avert more disability-adjusted life years (DALYs) per socioeconomic cost (measured by productive hours lost). Our results suggest any DCT method can help save lives, support re-opening of economies, and prevent second-wave outbreaks, and that FCT methods are a promising direction for enriching BCT using self-reported symptoms, yielding earlier warning signals and a significantly reduced spread of the virus per socioeconomic cost.
Predicting Infectiousness for Proactive Contact Tracing
Oct 23, 2020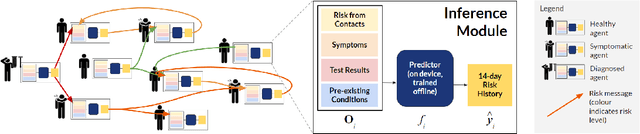
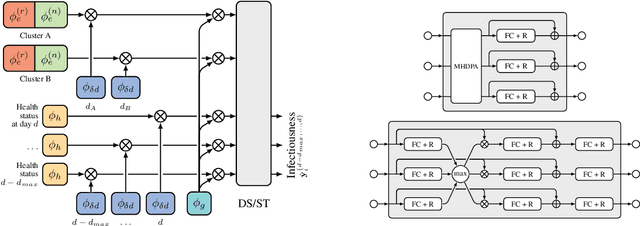
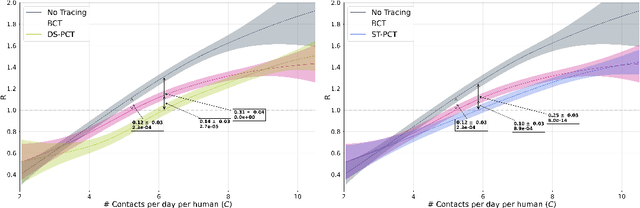
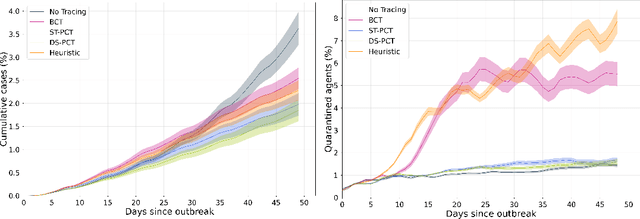
Abstract:The COVID-19 pandemic has spread rapidly worldwide, overwhelming manual contact tracing in many countries and resulting in widespread lockdowns for emergency containment. Large-scale digital contact tracing (DCT) has emerged as a potential solution to resume economic and social activity while minimizing spread of the virus. Various DCT methods have been proposed, each making trade-offs between privacy, mobility restrictions, and public health. The most common approach, binary contact tracing (BCT), models infection as a binary event, informed only by an individual's test results, with corresponding binary recommendations that either all or none of the individual's contacts quarantine. BCT ignores the inherent uncertainty in contacts and the infection process, which could be used to tailor messaging to high-risk individuals, and prompt proactive testing or earlier warnings. It also does not make use of observations such as symptoms or pre-existing medical conditions, which could be used to make more accurate infectiousness predictions. In this paper, we use a recently-proposed COVID-19 epidemiological simulator to develop and test methods that can be deployed to a smartphone to locally and proactively predict an individual's infectiousness (risk of infecting others) based on their contact history and other information, while respecting strong privacy constraints. Predictions are used to provide personalized recommendations to the individual via an app, as well as to send anonymized messages to the individual's contacts, who use this information to better predict their own infectiousness, an approach we call proactive contact tracing (PCT). We find a deep-learning based PCT method which improves over BCT for equivalent average mobility, suggesting PCT could help in safe re-opening and second-wave prevention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge