Aihua Liu
Methodology and Real-World Applications of Dynamic Uncertain Causality Graph for Clinical Diagnosis with Explainability and Invariance
Jun 09, 2024Abstract:AI-aided clinical diagnosis is desired in medical care. Existing deep learning models lack explainability and mainly focus on image analysis. The recently developed Dynamic Uncertain Causality Graph (DUCG) approach is causality-driven, explainable, and invariant across different application scenarios, without problems of data collection, labeling, fitting, privacy, bias, generalization, high cost and high energy consumption. Through close collaboration between clinical experts and DUCG technicians, 46 DUCG models covering 54 chief complaints were constructed. Over 1,000 diseases can be diagnosed without triage. Before being applied in real-world, the 46 DUCG models were retrospectively verified by third-party hospitals. The verified diagnostic precisions were no less than 95%, in which the diagnostic precision for every disease including uncommon ones was no less than 80%. After verifications, the 46 DUCG models were applied in the real-world in China. Over one million real diagnosis cases have been performed, with only 17 incorrect diagnoses identified. Due to DUCG's transparency, the mistakes causing the incorrect diagnoses were found and corrected. The diagnostic abilities of the clinicians who applied DUCG frequently were improved significantly. Following the introduction to the earlier presented DUCG methodology, the recommendation algorithm for potential medical checks is presented and the key idea of DUCG is extracted.
Supervised multi-specialist topic model with applications on large-scale electronic health record data
May 04, 2021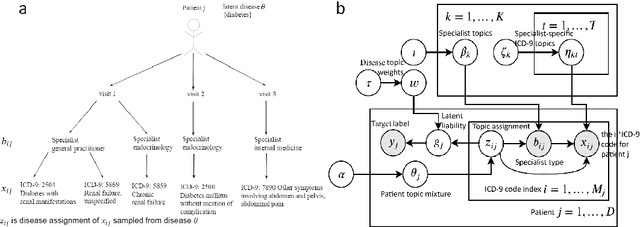
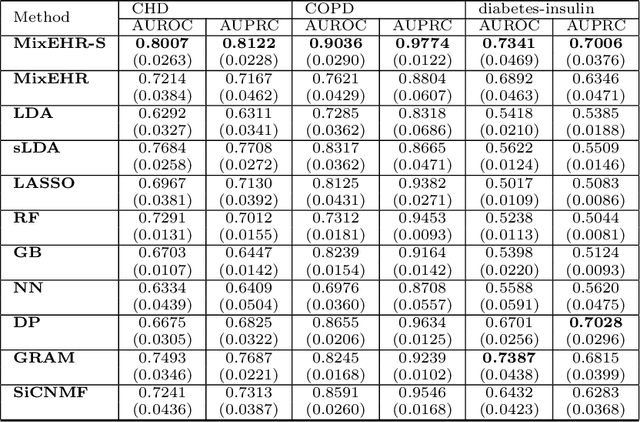
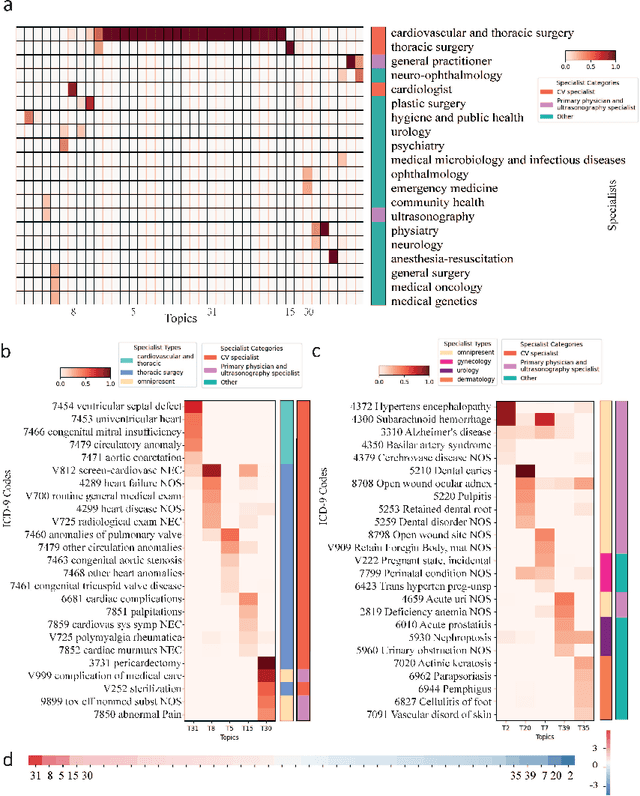
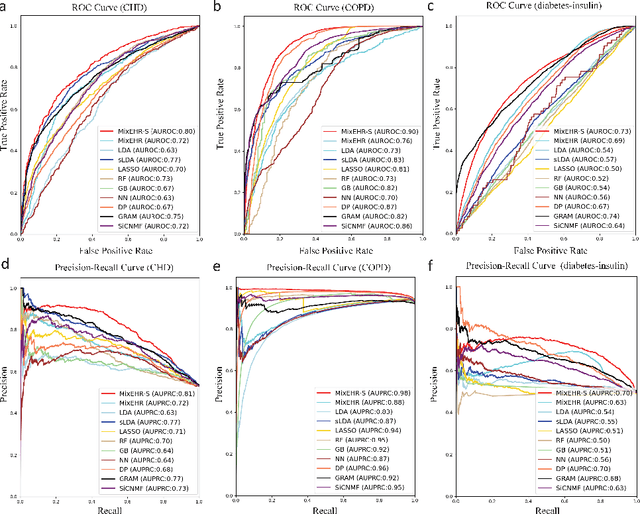
Abstract:Motivation: Electronic health record (EHR) data provides a new venue to elucidate disease comorbidities and latent phenotypes for precision medicine. To fully exploit its potential, a realistic data generative process of the EHR data needs to be modelled. We present MixEHR-S to jointly infer specialist-disease topics from the EHR data. As the key contribution, we model the specialist assignments and ICD-coded diagnoses as the latent topics based on patient's underlying disease topic mixture in a novel unified supervised hierarchical Bayesian topic model. For efficient inference, we developed a closed-form collapsed variational inference algorithm to learn the model distributions of MixEHR-S. We applied MixEHR-S to two independent large-scale EHR databases in Quebec with three targeted applications: (1) Congenital Heart Disease (CHD) diagnostic prediction among 154,775 patients; (2) Chronic obstructive pulmonary disease (COPD) diagnostic prediction among 73,791 patients; (3) future insulin treatment prediction among 78,712 patients diagnosed with diabetes as a mean to assess the disease exacerbation. In all three applications, MixEHR-S conferred clinically meaningful latent topics among the most predictive latent topics and achieved superior target prediction accuracy compared to the existing methods, providing opportunities for prioritizing high-risk patients for healthcare services. MixEHR-S source code and scripts of the experiments are freely available at https://github.com/li-lab-mcgill/mixehrS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge