Ariane Marelli
MixEHR-SurG: a joint proportional hazard and guided topic model for inferring mortality-associated topics from electronic health records
Dec 20, 2023Abstract:Objective: To improve survival analysis using EHR data, we aim to develop a supervised topic model called MixEHR-SurG to simultaneously integrate heterogeneous EHR data and model survival hazard. Materials and Methods: Our technical contributions are three-folds: (1) integrating EHR topic inference with Cox proportional hazards likelihood; (2) inferring patient-specific topic hyperparameters using the PheCode concepts such that each topic can be identified with exactly one PheCode-associated phenotype; (3) multi-modal survival topic inference. This leads to a highly interpretable survival and guided topic model that can infer PheCode-specific phenotype topics associated with patient mortality. We evaluated MixEHR-G using a simulated dataset and two real-world EHR datasets: the Quebec Congenital Heart Disease (CHD) data consisting of 8,211 subjects with 75,187 outpatient claim data of 1,767 unique ICD codes; the MIMIC-III consisting of 1,458 subjects with multi-modal EHR records. Results: Compared to the baselines, MixEHR-G achieved a superior dynamic AUROC for mortality prediction, with a mean AUROC score of 0.89 in the simulation dataset and a mean AUROC of 0.645 on the CHD dataset. Qualitatively, MixEHR-G associates severe cardiac conditions with high mortality risk among the CHD patients after the first heart failure hospitalization and critical brain injuries with increased mortality among the MIMIC-III patients after their ICU discharge. Conclusion: The integration of the Cox proportional hazards model and EHR topic inference in MixEHR-SurG led to not only competitive mortality prediction but also meaningful phenotype topics for systematic survival analysis. The software is available at GitHub: https://github.com/li-lab-mcgill/MixEHR-SurG.
Supervised multi-specialist topic model with applications on large-scale electronic health record data
May 04, 2021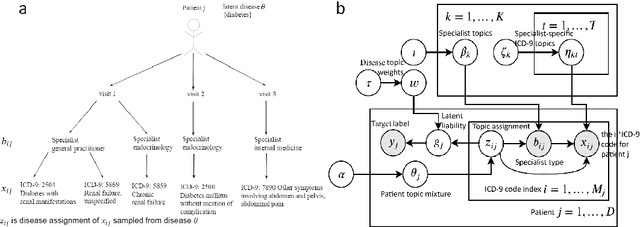
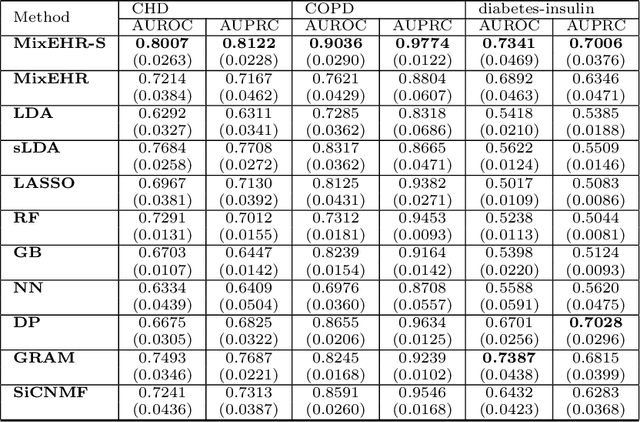
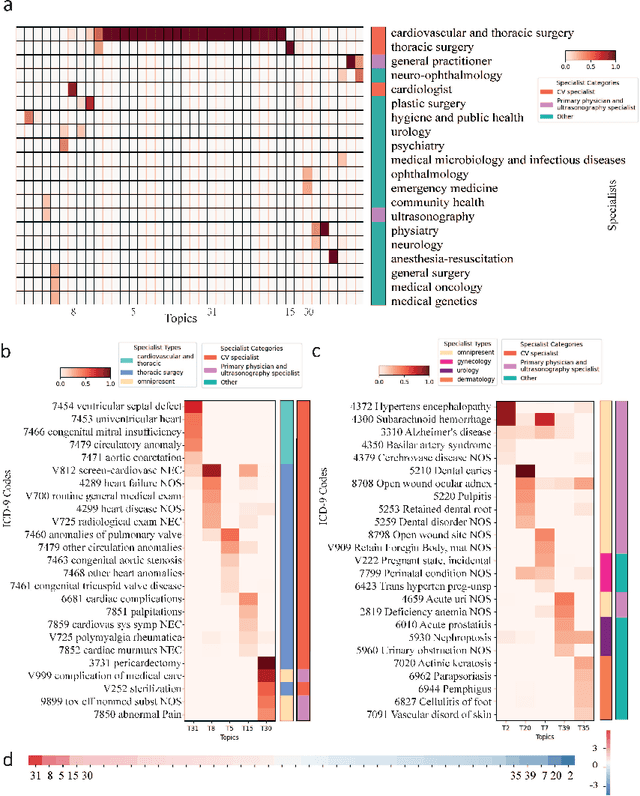
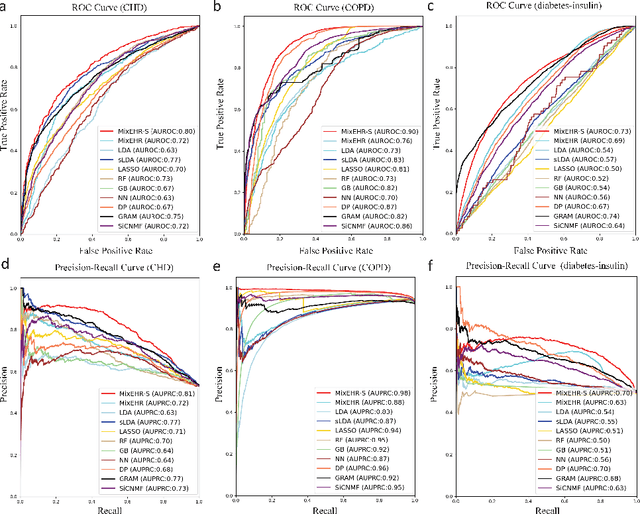
Abstract:Motivation: Electronic health record (EHR) data provides a new venue to elucidate disease comorbidities and latent phenotypes for precision medicine. To fully exploit its potential, a realistic data generative process of the EHR data needs to be modelled. We present MixEHR-S to jointly infer specialist-disease topics from the EHR data. As the key contribution, we model the specialist assignments and ICD-coded diagnoses as the latent topics based on patient's underlying disease topic mixture in a novel unified supervised hierarchical Bayesian topic model. For efficient inference, we developed a closed-form collapsed variational inference algorithm to learn the model distributions of MixEHR-S. We applied MixEHR-S to two independent large-scale EHR databases in Quebec with three targeted applications: (1) Congenital Heart Disease (CHD) diagnostic prediction among 154,775 patients; (2) Chronic obstructive pulmonary disease (COPD) diagnostic prediction among 73,791 patients; (3) future insulin treatment prediction among 78,712 patients diagnosed with diabetes as a mean to assess the disease exacerbation. In all three applications, MixEHR-S conferred clinically meaningful latent topics among the most predictive latent topics and achieved superior target prediction accuracy compared to the existing methods, providing opportunities for prioritizing high-risk patients for healthcare services. MixEHR-S source code and scripts of the experiments are freely available at https://github.com/li-lab-mcgill/mixehrS
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge