Archer Y. Yang
Multivariate Conformal Selection
May 01, 2025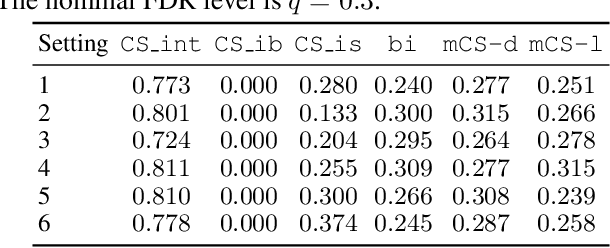
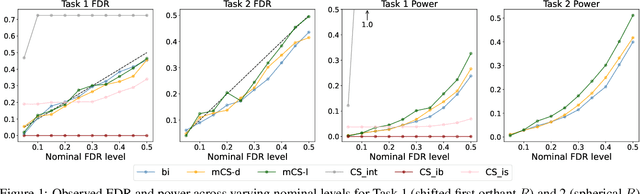
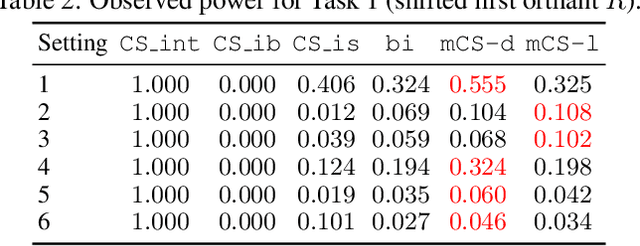
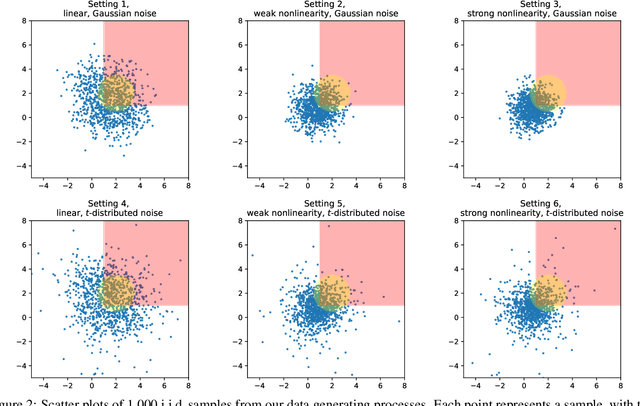
Abstract:Selecting high-quality candidates from large datasets is critical in applications such as drug discovery, precision medicine, and alignment of large language models (LLMs). While Conformal Selection (CS) provides rigorous uncertainty quantification, it is limited to univariate responses and scalar criteria. To address this issue, we propose Multivariate Conformal Selection (mCS), a generalization of CS designed for multivariate response settings. Our method introduces regional monotonicity and employs multivariate nonconformity scores to construct conformal p-values, enabling finite-sample False Discovery Rate (FDR) control. We present two variants: mCS-dist, using distance-based scores, and mCS-learn, which learns optimal scores via differentiable optimization. Experiments on simulated and real-world datasets demonstrate that mCS significantly improves selection power while maintaining FDR control, establishing it as a robust framework for multivariate selection tasks.
QComp: A QSAR-Based Data Completion Framework for Drug Discovery
May 20, 2024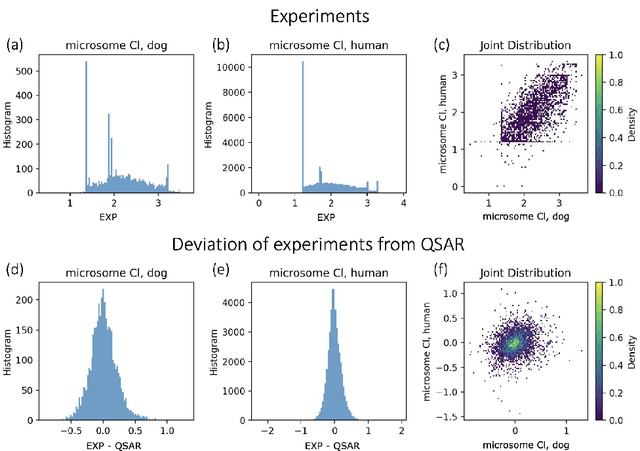
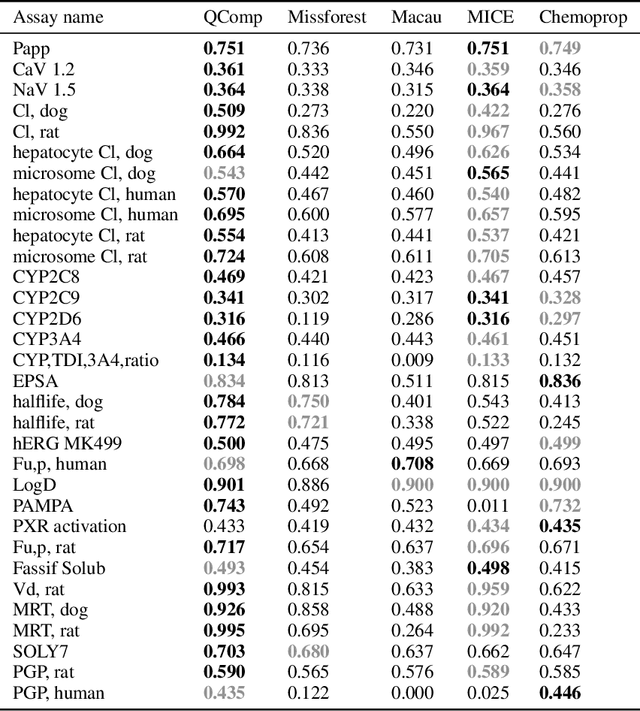
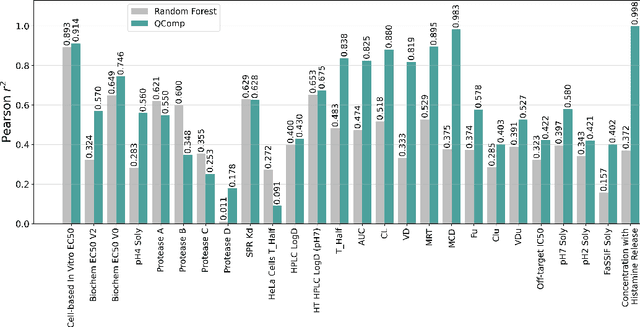
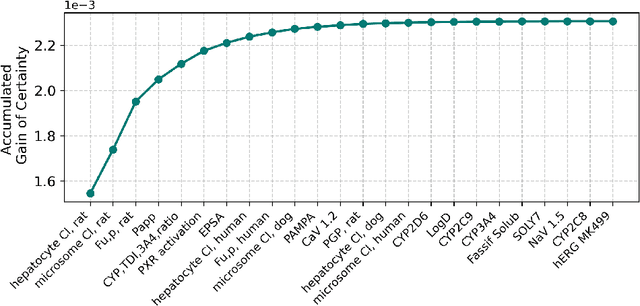
Abstract:In drug discovery, in vitro and in vivo experiments reveal biochemical activities related to the efficacy and toxicity of compounds. The experimental data accumulate into massive, ever-evolving, and sparse datasets. Quantitative Structure-Activity Relationship (QSAR) models, which predict biochemical activities using only the structural information of compounds, face challenges in integrating the evolving experimental data as studies progress. We develop QSAR-Complete (QComp), a data completion framework to address this issue. Based on pre-existing QSAR models, QComp utilizes the correlation inherent in experimental data to enhance prediction accuracy across various tasks. Moreover, QComp emerges as a promising tool for guiding the optimal sequence of experiments by quantifying the reduction in statistical uncertainty for specific endpoints, thereby aiding in rational decision-making throughout the drug discovery process.
MixEHR-SurG: a joint proportional hazard and guided topic model for inferring mortality-associated topics from electronic health records
Dec 20, 2023Abstract:Objective: To improve survival analysis using EHR data, we aim to develop a supervised topic model called MixEHR-SurG to simultaneously integrate heterogeneous EHR data and model survival hazard. Materials and Methods: Our technical contributions are three-folds: (1) integrating EHR topic inference with Cox proportional hazards likelihood; (2) inferring patient-specific topic hyperparameters using the PheCode concepts such that each topic can be identified with exactly one PheCode-associated phenotype; (3) multi-modal survival topic inference. This leads to a highly interpretable survival and guided topic model that can infer PheCode-specific phenotype topics associated with patient mortality. We evaluated MixEHR-G using a simulated dataset and two real-world EHR datasets: the Quebec Congenital Heart Disease (CHD) data consisting of 8,211 subjects with 75,187 outpatient claim data of 1,767 unique ICD codes; the MIMIC-III consisting of 1,458 subjects with multi-modal EHR records. Results: Compared to the baselines, MixEHR-G achieved a superior dynamic AUROC for mortality prediction, with a mean AUROC score of 0.89 in the simulation dataset and a mean AUROC of 0.645 on the CHD dataset. Qualitatively, MixEHR-G associates severe cardiac conditions with high mortality risk among the CHD patients after the first heart failure hospitalization and critical brain injuries with increased mortality among the MIMIC-III patients after their ICU discharge. Conclusion: The integration of the Cox proportional hazards model and EHR topic inference in MixEHR-SurG led to not only competitive mortality prediction but also meaningful phenotype topics for systematic survival analysis. The software is available at GitHub: https://github.com/li-lab-mcgill/MixEHR-SurG.
Structured Learning in Time-dependent Cox Models
Jun 21, 2023Abstract:Cox models with time-dependent coefficients and covariates are widely used in survival analysis. In high-dimensional settings, sparse regularization techniques are employed for variable selection, but existing methods for time-dependent Cox models lack flexibility in enforcing specific sparsity patterns (i.e., covariate structures). We propose a flexible framework for variable selection in time-dependent Cox models, accommodating complex selection rules. Our method can adapt to arbitrary grouping structures, including interaction selection, temporal, spatial, tree, and directed acyclic graph structures. It achieves accurate estimation with low false alarm rates. We develop the sox package, implementing a network flow algorithm for efficiently solving models with complex covariate structures. Sox offers a user-friendly interface for specifying grouping structures and delivers fast computation. Through examples, including a case study on identifying predictors of time to all-cause death in atrial fibrillation patients, we demonstrate the practical application of our method with specific selection rules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge