Byung-Jun Yoon
C-LoRA: Contextual Low-Rank Adaptation for Uncertainty Estimation in Large Language Models
May 23, 2025Abstract:Low-Rank Adaptation (LoRA) offers a cost-effective solution for fine-tuning large language models (LLMs), but it often produces overconfident predictions in data-scarce few-shot settings. To address this issue, several classical statistical learning approaches have been repurposed for scalable uncertainty-aware LoRA fine-tuning. However, these approaches neglect how input characteristics affect the predictive uncertainty estimates. To address this limitation, we propose Contextual Low-Rank Adaptation (\textbf{C-LoRA}) as a novel uncertainty-aware and parameter efficient fine-tuning approach, by developing new lightweight LoRA modules contextualized to each input data sample to dynamically adapt uncertainty estimates. Incorporating data-driven contexts into the parameter posteriors, C-LoRA mitigates overfitting, achieves well-calibrated uncertainties, and yields robust predictions. Extensive experiments demonstrate that C-LoRA consistently outperforms the state-of-the-art uncertainty-aware LoRA methods in both uncertainty quantification and model generalization. Ablation studies further confirm the critical role of our contextual modules in capturing sample-specific uncertainties. C-LoRA sets a new standard for robust, uncertainty-aware LLM fine-tuning in few-shot regimes.
Uncertainty-Aware Adaptation of Large Language Models for Protein-Protein Interaction Analysis
Feb 10, 2025



Abstract:Identification of protein-protein interactions (PPIs) helps derive cellular mechanistic understanding, particularly in the context of complex conditions such as neurodegenerative disorders, metabolic syndromes, and cancer. Large Language Models (LLMs) have demonstrated remarkable potential in predicting protein structures and interactions via automated mining of vast biomedical literature; yet their inherent uncertainty remains a key challenge for deriving reproducible findings, critical for biomedical applications. In this study, we present an uncertainty-aware adaptation of LLMs for PPI analysis, leveraging fine-tuned LLaMA-3 and BioMedGPT models. To enhance prediction reliability, we integrate LoRA ensembles and Bayesian LoRA models for uncertainty quantification (UQ), ensuring confidence-calibrated insights into protein behavior. Our approach achieves competitive performance in PPI identification across diverse disease contexts while addressing model uncertainty, thereby enhancing trustworthiness and reproducibility in computational biology. These findings underscore the potential of uncertainty-aware LLM adaptation for advancing precision medicine and biomedical research.
Epidemiological Model Calibration via Graybox Bayesian Optimization
Dec 10, 2024



Abstract:In this study, we focus on developing efficient calibration methods via Bayesian decision-making for the family of compartmental epidemiological models. The existing calibration methods usually assume that the compartmental model is cheap in terms of its output and gradient evaluation, which may not hold in practice when extending them to more general settings. Therefore, we introduce model calibration methods based on a "graybox" Bayesian optimization (BO) scheme, more efficient calibration for general epidemiological models. This approach uses Gaussian processes as a surrogate to the expensive model, and leverages the functional structure of the compartmental model to enhance calibration performance. Additionally, we develop model calibration methods via a decoupled decision-making strategy for BO, which further exploits the decomposable nature of the functional structure. The calibration efficiencies of the multiple proposed schemes are evaluated based on various data generated by a compartmental model mimicking real-world epidemic processes, and real-world COVID-19 datasets. Experimental results demonstrate that our proposed graybox variants of BO schemes can efficiently calibrate computationally expensive models and further improve the calibration performance measured by the logarithm of mean square errors and achieve faster performance convergence in terms of BO iterations. We anticipate that the proposed calibration methods can be extended to enable fast calibration of more complex epidemiological models, such as the agent-based models.
Efficient Compression of Sparse Accelerator Data Using Implicit Neural Representations and Importance Sampling
Dec 02, 2024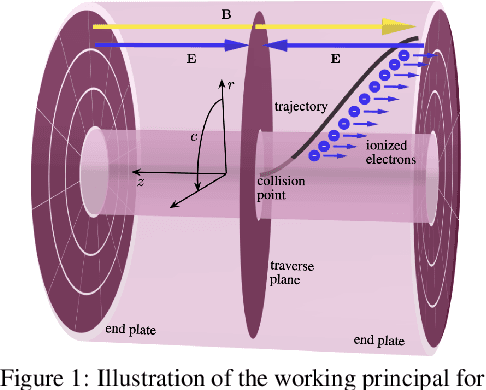
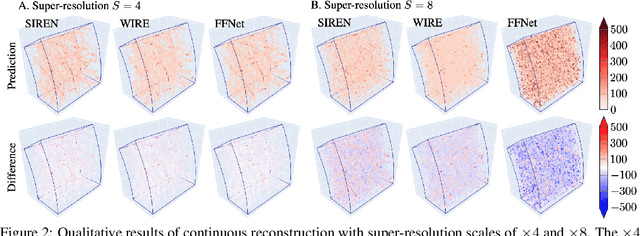
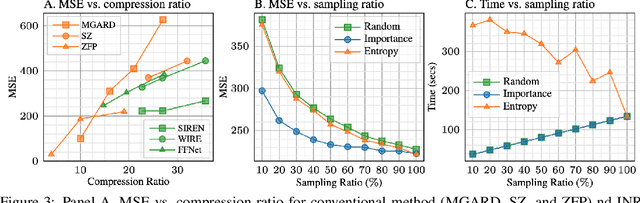
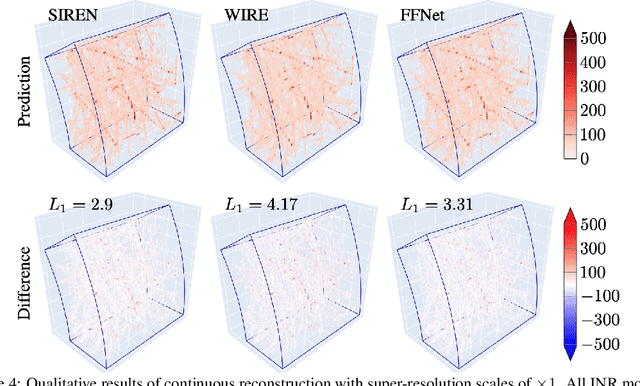
Abstract:High-energy, large-scale particle colliders in nuclear and high-energy physics generate data at extraordinary rates, reaching up to $1$ terabyte and several petabytes per second, respectively. The development of real-time, high-throughput data compression algorithms capable of reducing this data to manageable sizes for permanent storage is of paramount importance. A unique characteristic of the tracking detector data is the extreme sparsity of particle trajectories in space, with an occupancy rate ranging from approximately $10^{-6}$ to $10\%$. Furthermore, for downstream tasks, a continuous representation of this data is often more useful than a voxel-based, discrete representation due to the inherently continuous nature of the signals involved. To address these challenges, we propose a novel approach using implicit neural representations for data learning and compression. We also introduce an importance sampling technique to accelerate the network training process. Our method is competitive with traditional compression algorithms, such as MGARD, SZ, and ZFP, while offering significant speed-ups and maintaining negligible accuracy loss through our importance sampling strategy.
Variable Rate Neural Compression for Sparse Detector Data
Nov 18, 2024Abstract:High-energy large-scale particle colliders generate data at extraordinary rates. Developing real-time high-throughput data compression algorithms to reduce data volume and meet the bandwidth requirement for storage has become increasingly critical. Deep learning is a promising technology that can address this challenging topic. At the newly constructed sPHENIX experiment at the Relativistic Heavy Ion Collider, a Time Projection Chamber (TPC) serves as the main tracking detector, which records three-dimensional particle trajectories in a volume of a gas-filled cylinder. In terms of occupancy, the resulting data flow can be very sparse reaching $10^{-3}$ for proton-proton collisions. Such sparsity presents a challenge to conventional learning-free lossy compression algorithms, such as SZ, ZFP, and MGARD. In contrast, emerging deep learning-based models, particularly those utilizing convolutional neural networks for compression, have outperformed these conventional methods in terms of compression ratios and reconstruction accuracy. However, research on the efficacy of these deep learning models in handling sparse datasets, like those produced in particle colliders, remains limited. Furthermore, most deep learning models do not adapt their processing speeds to data sparsity, which affects efficiency. To address this issue, we propose a novel approach for TPC data compression via key-point identification facilitated by sparse convolution. Our proposed algorithm, BCAE-VS, achieves a $75\%$ improvement in reconstruction accuracy with a $10\%$ increase in compression ratio over the previous state-of-the-art model. Additionally, BCAE-VS manages to achieve these results with a model size over two orders of magnitude smaller. Lastly, we have experimentally verified that as sparsity increases, so does the model's throughput.
LoRA-BERT: a Natural Language Processing Model for Robust and Accurate Prediction of long non-coding RNAs
Nov 11, 2024Abstract:Long non-coding RNAs (lncRNAs) serve as crucial regulators in numerous biological processes. Although they share sequence similarities with messenger RNAs (mRNAs), lncRNAs perform entirely different roles, providing new avenues for biological research. The emergence of next-generation sequencing technologies has greatly advanced the detection and identification of lncRNA transcripts and deep learning-based approaches have been introduced to classify long non-coding RNAs (lncRNAs). These advanced methods have significantly enhanced the efficiency of identifying lncRNAs. However, many of these methods are devoid of robustness and accuracy due to the extended length of the sequences involved. To tackle this issue, we have introduced a novel pre-trained bidirectional encoder representation called LoRA-BERT. LoRA-BERT is designed to capture the importance of nucleotide-level information during sequence classification, leading to more robust and satisfactory outcomes. In a comprehensive comparison with commonly used sequence prediction tools, we have demonstrated that LoRA-BERT outperforms them in terms of accuracy and efficiency. Our results indicate that, when utilizing the transformer model, LoRA-BERT achieves state-of-the-art performance in predicting both lncRNAs and mRNAs for human and mouse species. Through the utilization of LoRA-BERT, we acquire valuable insights into the traits of lncRNAs and mRNAs, offering the potential to aid in the comprehension and detection of diseases linked to lncRNAs in humans.
Pathway-Guided Optimization of Deep Generative Molecular Design Models for Cancer Therapy
Nov 05, 2024



Abstract:The data-driven drug design problem can be formulated as an optimization task of a potentially expensive black-box objective function over a huge high-dimensional and structured molecular space. The junction tree variational autoencoder (JTVAE) has been shown to be an efficient generative model that can be used for suggesting legitimate novel drug-like small molecules with improved properties. While the performance of the generative molecular design (GMD) scheme strongly depends on the initial training data, one can improve its sampling efficiency for suggesting better molecules with enhanced properties by optimizing the latent space. In this work, we propose how mechanistic models - such as pathway models described by differential equations - can be used for effective latent space optimization(LSO) of JTVAEs and other similar models for GMD. To demonstrate the potential of our proposed approach, we show how a pharmacodynamic model, assessing the therapeutic efficacy of a drug-like small molecule by predicting how it modulates a cancer pathway, can be incorporated for effective LSO of data-driven models for GMD.
Enhancing Future Link Prediction in Quantum Computing Semantic Networks through LLM-Initiated Node Features
Oct 05, 2024



Abstract:Quantum computing is rapidly evolving in both physics and computer science, offering the potential to solve complex problems and accelerate computational processes. The development of quantum chips necessitates understanding the correlations among diverse experimental conditions. Semantic networks built on scientific literature, representing meaningful relationships between concepts, have been used across various domains to identify knowledge gaps and novel concept combinations. Neural network-based approaches have shown promise in link prediction within these networks. This study proposes initializing node features using LLMs to enhance node representations for link prediction tasks in graph neural networks. LLMs can provide rich descriptions, reducing the need for manual feature creation and lowering costs. Our method, evaluated using various link prediction models on a quantum computing semantic network, demonstrated efficacy compared to traditional node embedding techniques.
Understanding Uncertainty-based Active Learning Under Model Mismatch
Aug 24, 2024



Abstract:Instead of randomly acquiring training data points, Uncertainty-based Active Learning (UAL) operates by querying the label(s) of pivotal samples from an unlabeled pool selected based on the prediction uncertainty, thereby aiming at minimizing the labeling cost for model training. The efficacy of UAL critically depends on the model capacity as well as the adopted uncertainty-based acquisition function. Within the context of this study, our analytical focus is directed toward comprehending how the capacity of the machine learning model may affect UAL efficacy. Through theoretical analysis, comprehensive simulations, and empirical studies, we conclusively demonstrate that UAL can lead to worse performance in comparison with random sampling when the machine learning model class has low capacity and is unable to cover the underlying ground truth. In such situations, adopting acquisition functions that directly target estimating the prediction performance may be beneficial for improving the performance of UAL.
Enhancing Generative Molecular Design via Uncertainty-guided Fine-tuning of Variational Autoencoders
May 31, 2024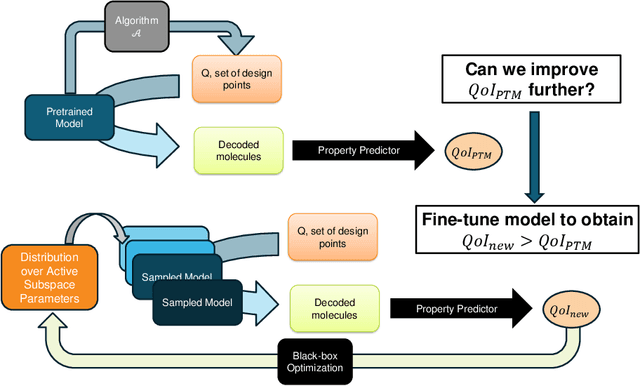
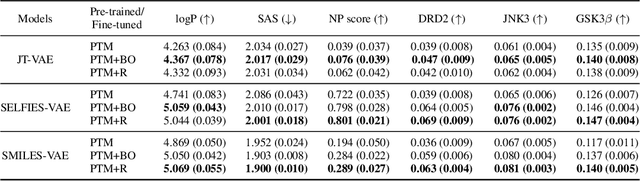
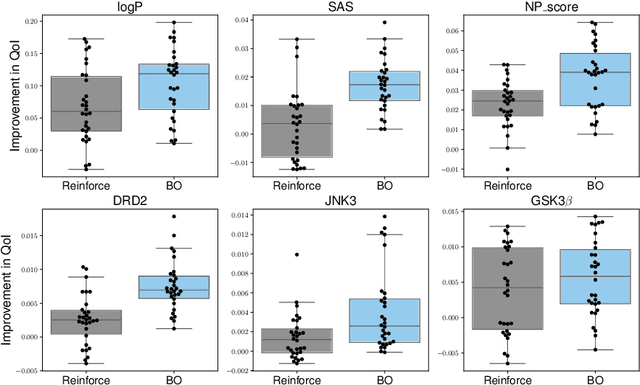
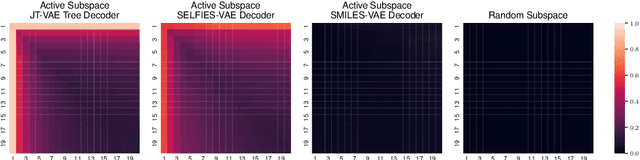
Abstract:In recent years, deep generative models have been successfully adopted for various molecular design tasks, particularly in the life and material sciences. A critical challenge for pre-trained generative molecular design (GMD) models is to fine-tune them to be better suited for downstream design tasks aimed at optimizing specific molecular properties. However, redesigning and training an existing effective generative model from scratch for each new design task is impractical. Furthermore, the black-box nature of typical downstream tasks$\unicode{x2013}$such as property prediction$\unicode{x2013}$makes it nontrivial to optimize the generative model in a task-specific manner. In this work, we propose a novel approach for a model uncertainty-guided fine-tuning of a pre-trained variational autoencoder (VAE)-based GMD model through performance feedback in an active learning setting. The main idea is to quantify model uncertainty in the generative model, which is made efficient by working within a low-dimensional active subspace of the high-dimensional VAE parameters explaining most of the variability in the model's output. The inclusion of model uncertainty expands the space of viable molecules through decoder diversity. We then explore the resulting model uncertainty class via black-box optimization made tractable by low-dimensionality of the active subspace. This enables us to identify and leverage a diverse set of high-performing models to generate enhanced molecules. Empirical results across six target molecular properties, using multiple VAE-based generative models, demonstrate that our uncertainty-guided fine-tuning approach consistently outperforms the original pre-trained models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge