Gilchan Park
Uncertainty-Aware Adaptation of Large Language Models for Protein-Protein Interaction Analysis
Feb 10, 2025



Abstract:Identification of protein-protein interactions (PPIs) helps derive cellular mechanistic understanding, particularly in the context of complex conditions such as neurodegenerative disorders, metabolic syndromes, and cancer. Large Language Models (LLMs) have demonstrated remarkable potential in predicting protein structures and interactions via automated mining of vast biomedical literature; yet their inherent uncertainty remains a key challenge for deriving reproducible findings, critical for biomedical applications. In this study, we present an uncertainty-aware adaptation of LLMs for PPI analysis, leveraging fine-tuned LLaMA-3 and BioMedGPT models. To enhance prediction reliability, we integrate LoRA ensembles and Bayesian LoRA models for uncertainty quantification (UQ), ensuring confidence-calibrated insights into protein behavior. Our approach achieves competitive performance in PPI identification across diverse disease contexts while addressing model uncertainty, thereby enhancing trustworthiness and reproducibility in computational biology. These findings underscore the potential of uncertainty-aware LLM adaptation for advancing precision medicine and biomedical research.
Enhancing Future Link Prediction in Quantum Computing Semantic Networks through LLM-Initiated Node Features
Oct 05, 2024



Abstract:Quantum computing is rapidly evolving in both physics and computer science, offering the potential to solve complex problems and accelerate computational processes. The development of quantum chips necessitates understanding the correlations among diverse experimental conditions. Semantic networks built on scientific literature, representing meaningful relationships between concepts, have been used across various domains to identify knowledge gaps and novel concept combinations. Neural network-based approaches have shown promise in link prediction within these networks. This study proposes initializing node features using LLMs to enhance node representations for link prediction tasks in graph neural networks. LLMs can provide rich descriptions, reducing the need for manual feature creation and lowering costs. Our method, evaluated using various link prediction models on a quantum computing semantic network, demonstrated efficacy compared to traditional node embedding techniques.
Extracting Protein-Protein Interactions (PPIs) from Biomedical Literature using Attention-based Relational Context Information
Mar 08, 2024Abstract:Because protein-protein interactions (PPIs) are crucial to understand living systems, harvesting these data is essential to probe disease development and discern gene/protein functions and biological processes. Some curated datasets contain PPI data derived from the literature and other sources (e.g., IntAct, BioGrid, DIP, and HPRD). However, they are far from exhaustive, and their maintenance is a labor-intensive process. On the other hand, machine learning methods to automate PPI knowledge extraction from the scientific literature have been limited by a shortage of appropriate annotated data. This work presents a unified, multi-source PPI corpora with vetted interaction definitions augmented by binary interaction type labels and a Transformer-based deep learning method that exploits entities' relational context information for relation representation to improve relation classification performance. The model's performance is evaluated on four widely studied biomedical relation extraction datasets, as well as this work's target PPI datasets, to observe the effectiveness of the representation to relation extraction tasks in various data. Results show the model outperforms prior state-of-the-art models. The code and data are available at: https://github.com/BNLNLP/PPI-Relation-Extraction
* 10 pages, 3 figures, 7 tables, 2022 IEEE International Conference on Big Data (Big Data)
Density Estimation via Measure Transport: Outlook for Applications in the Biological Sciences
Sep 27, 2023Abstract:One among several advantages of measure transport methods is that they allow for a unified framework for processing and analysis of data distributed according to a wide class of probability measures. Within this context, we present results from computational studies aimed at assessing the potential of measure transport techniques, specifically, the use of triangular transport maps, as part of a workflow intended to support research in the biological sciences. Scarce data scenarios, which are common in domains such as radiation biology, are of particular interest. We find that when data is scarce, sparse transport maps are advantageous. In particular, statistics gathered from computing series of (sparse) adaptive transport maps, trained on a series of randomly chosen subsets of the set of available data samples, leads to uncovering information hidden in the data. As a result, in the radiation biology application considered here, this approach provides a tool for generating hypotheses about gene relationships and their dynamics under radiation exposure.
Comparative Performance Evaluation of Large Language Models for Extracting Molecular Interactions and Pathway Knowledge
Jul 17, 2023



Abstract:Understanding protein interactions and pathway knowledge is crucial for unraveling the complexities of living systems and investigating the underlying mechanisms of biological functions and complex diseases. While existing databases provide curated biological data from literature and other sources, they are often incomplete and their maintenance is labor-intensive, necessitating alternative approaches. In this study, we propose to harness the capabilities of large language models to address these issues by automatically extracting such knowledge from the relevant scientific literature. Toward this goal, in this work, we investigate the effectiveness of different large language models in tasks that involve recognizing protein interactions, pathways, and gene regulatory relations. We thoroughly evaluate the performance of various models, highlight the significant findings, and discuss both the future opportunities and the remaining challenges associated with this approach. The code and data are available at: https://github.com/boxorange/BioIE-LLM
Comprehensive analysis of gene expression profiles to radiation exposure reveals molecular signatures of low-dose radiation response
Jan 03, 2023



Abstract:There are various sources of ionizing radiation exposure, where medical exposure for radiation therapy or diagnosis is the most common human-made source. Understanding how gene expression is modulated after ionizing radiation exposure and investigating the presence of any dose-dependent gene expression patterns have broad implications for health risks from radiotherapy, medical radiation diagnostic procedures, as well as other environmental exposure. In this paper, we perform a comprehensive pathway-based analysis of gene expression profiles in response to low-dose radiation exposure, in order to examine the potential mechanism of gene regulation underlying such responses. To accomplish this goal, we employ a statistical framework to determine whether a specific group of genes belonging to a known pathway display coordinated expression patterns that are modulated in a manner consistent with the radiation level. Findings in our study suggest that there exist complex yet consistent signatures that reflect the molecular response to radiation exposure, which differ between low-dose and high-dose radiation.
Figure Descriptive Text Extraction using Ontological Representation
Aug 11, 2022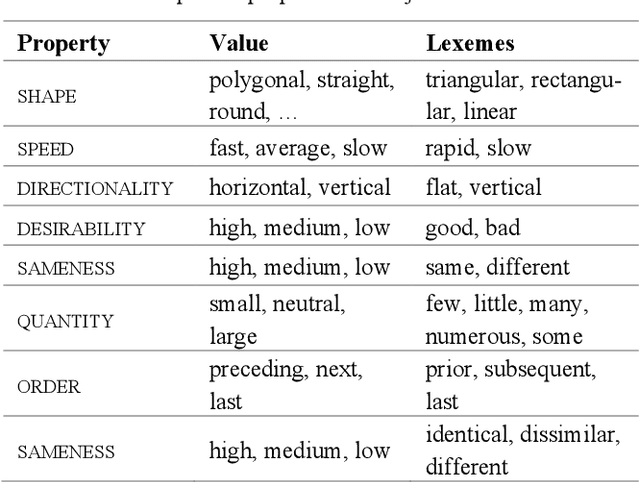
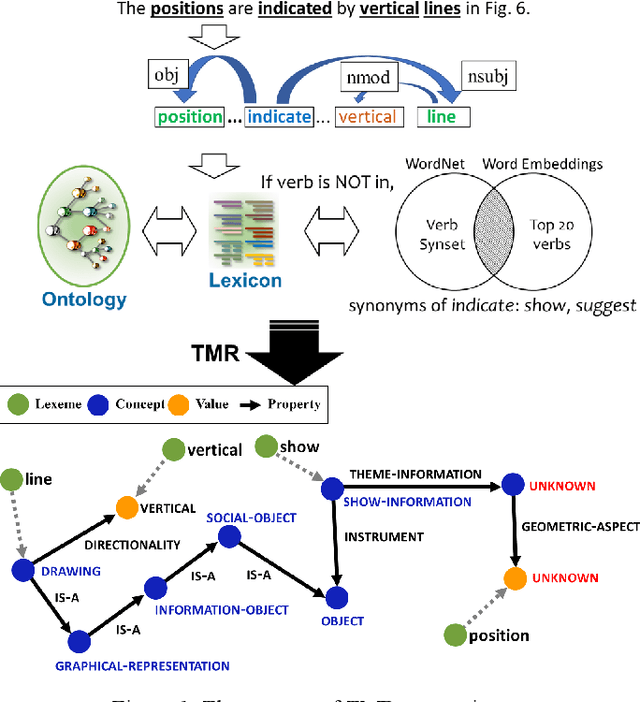
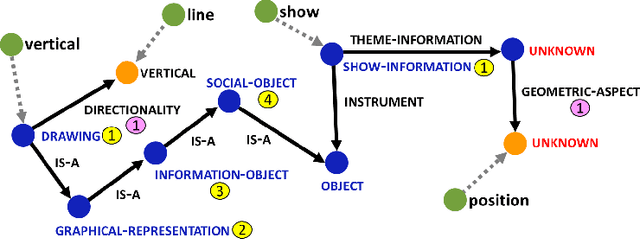
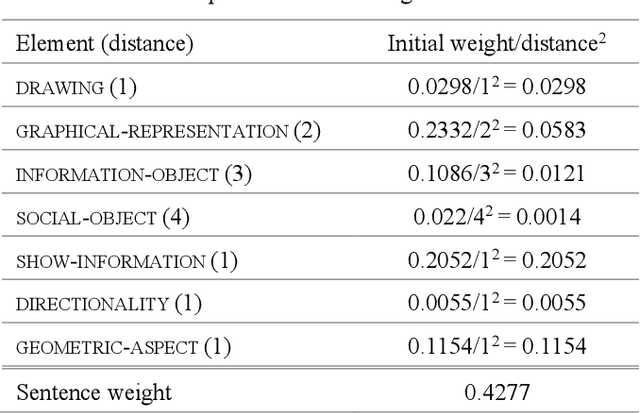
Abstract:Experimental research publications provide figure form resources including graphs, charts, and any type of images to effectively support and convey methods and results. To describe figures, authors add captions, which are often incomplete, and more descriptions reside in body text. This work presents a method to extract figure descriptive text from the body of scientific articles. We adopted ontological semantics to aid concept recognition of figure-related information, which generates human- and machine-readable knowledge representations from sentences. Our results show that conceptual models bring an improvement in figure descriptive sentence classification over word-based approaches.
Towards Text-based Phishing Detection
Nov 03, 2021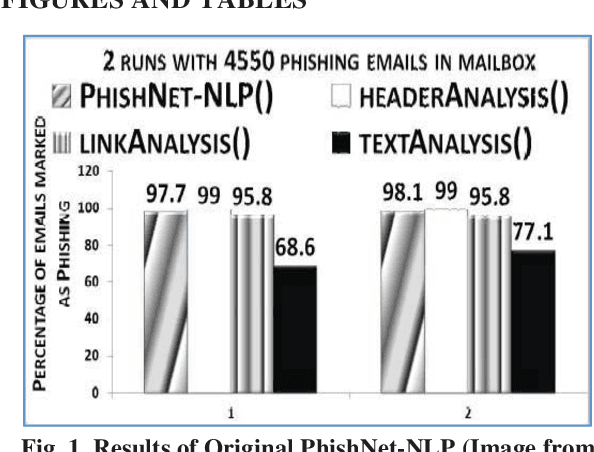
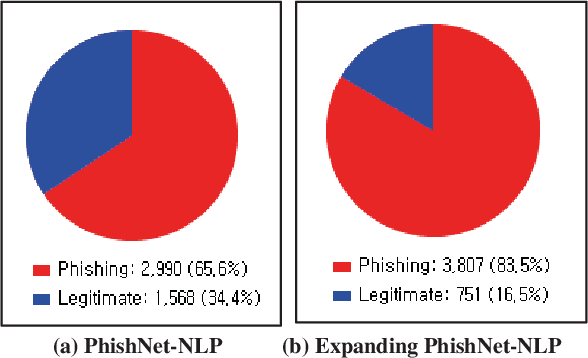
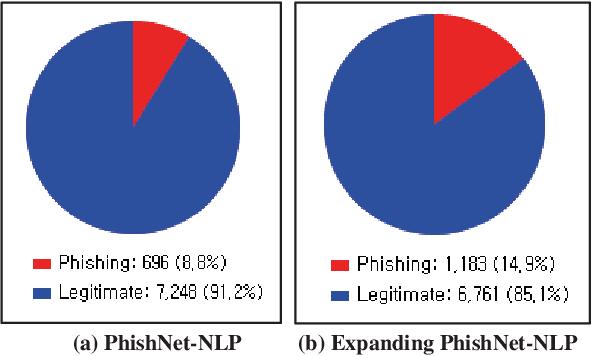
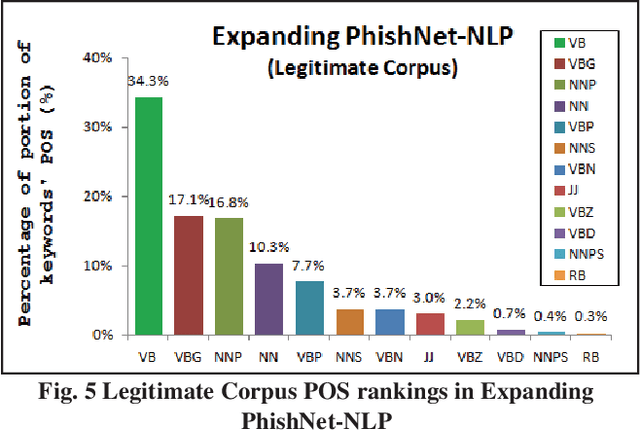
Abstract:This paper reports on an experiment into text-based phishing detection using readily available resources and without the use of semantics. The developed algorithm is a modified version of previously published work that works with the same tools. The results obtained in recognizing phishing emails are considerably better than the previously reported work; but the rate of text falsely identified as phishing is slightly worse. It is expected that adding semantic component will reduce the false positive rate while preserving the detection accuracy.
Towards The Automatic Coding of Medical Transcripts to Improve Patient-Centered Communication
Sep 22, 2021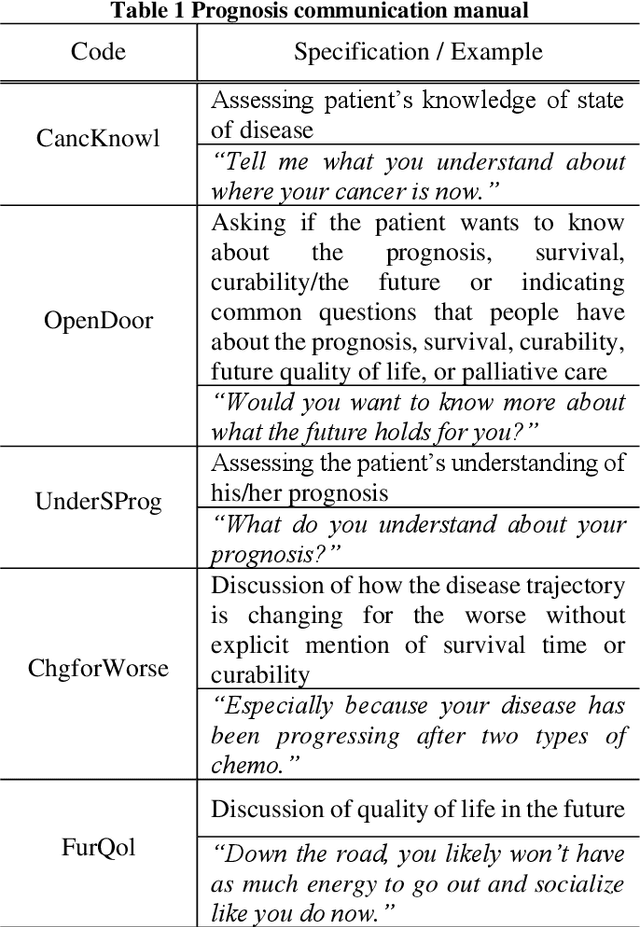
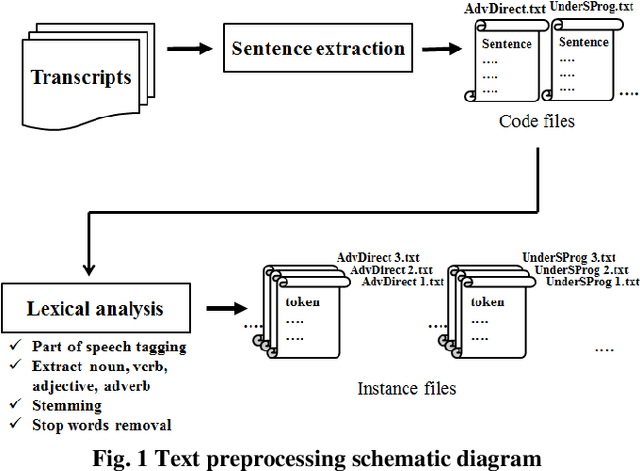
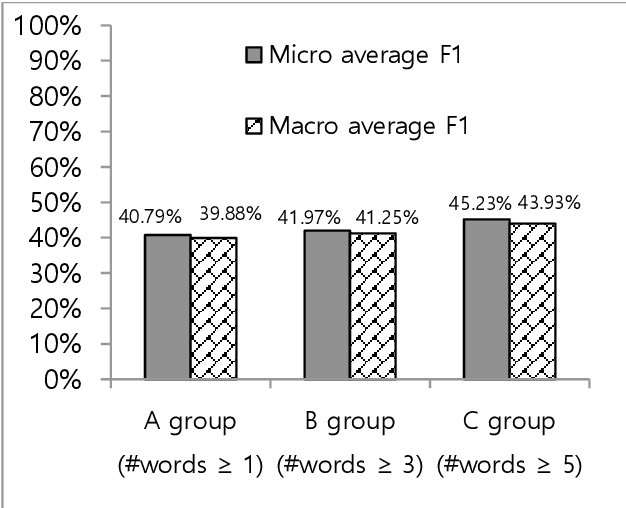
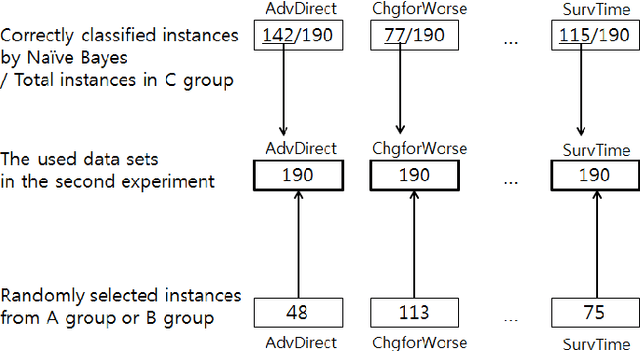
Abstract:This paper aims to provide an approach for automatic coding of physician-patient communication transcripts to improve patient-centered communication (PCC). PCC is a central part of high-quality health care. To improve PCC, dialogues between physicians and patients have been recorded and tagged with predefined codes. Trained human coders have manually coded the transcripts. Since it entails huge labor costs and poses possible human errors, automatic coding methods should be considered for efficiency and effectiveness. We adopted three machine learning algorithms (Na\"ive Bayes, Random Forest, and Support Vector Machine) to categorize lines in transcripts into corresponding codes. The result showed that there is evidence to distinguish the codes, and this is considered to be sufficient for training of human annotators.
Using Syntactic Features for Phishing Detection
May 29, 2015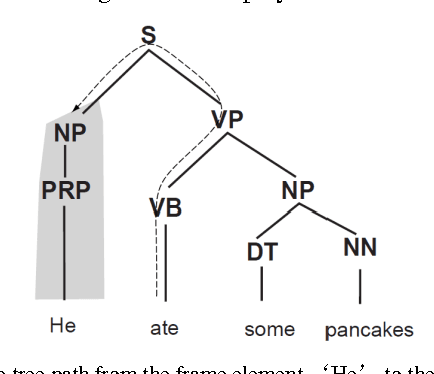

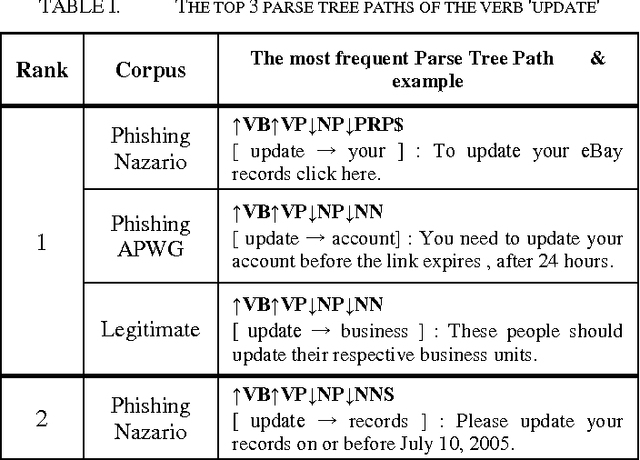
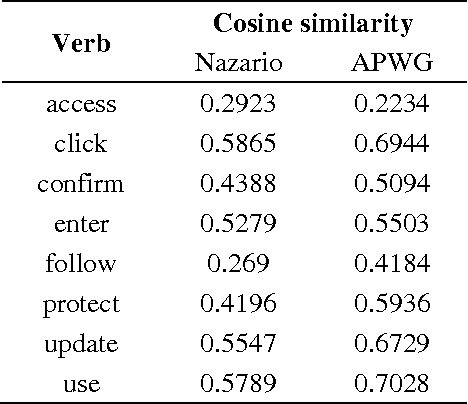
Abstract:This paper reports on the comparison of the subject and object of verbs in their usage between phishing emails and legitimate emails. The purpose of this research is to explore whether the syntactic structures and subjects and objects of verbs can be distinguishable features for phishing detection. To achieve the objective, we have conducted two series of experiments: the syntactic similarity for sentences, and the subject and object of verb comparison. The results of the experiments indicated that both features can be used for some verbs, but more work has to be done for others.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge