A N M Nafiz Abeer
Enhancing Generative Molecular Design via Uncertainty-guided Fine-tuning of Variational Autoencoders
May 31, 2024Abstract:In recent years, deep generative models have been successfully adopted for various molecular design tasks, particularly in the life and material sciences. A critical challenge for pre-trained generative molecular design (GMD) models is to fine-tune them to be better suited for downstream design tasks aimed at optimizing specific molecular properties. However, redesigning and training an existing effective generative model from scratch for each new design task is impractical. Furthermore, the black-box nature of typical downstream tasks$\unicode{x2013}$such as property prediction$\unicode{x2013}$makes it nontrivial to optimize the generative model in a task-specific manner. In this work, we propose a novel approach for a model uncertainty-guided fine-tuning of a pre-trained variational autoencoder (VAE)-based GMD model through performance feedback in an active learning setting. The main idea is to quantify model uncertainty in the generative model, which is made efficient by working within a low-dimensional active subspace of the high-dimensional VAE parameters explaining most of the variability in the model's output. The inclusion of model uncertainty expands the space of viable molecules through decoder diversity. We then explore the resulting model uncertainty class via black-box optimization made tractable by low-dimensionality of the active subspace. This enables us to identify and leverage a diverse set of high-performing models to generate enhanced molecules. Empirical results across six target molecular properties, using multiple VAE-based generative models, demonstrate that our uncertainty-guided fine-tuning approach consistently outperforms the original pre-trained models.
Leveraging Active Subspaces to Capture Epistemic Model Uncertainty in Deep Generative Models for Molecular Design
Apr 30, 2024Abstract:Deep generative models have been accelerating the inverse design process in material and drug design. Unlike their counterpart property predictors in typical molecular design frameworks, generative molecular design models have seen fewer efforts on uncertainty quantification (UQ) due to computational challenges in Bayesian inference posed by their large number of parameters. In this work, we focus on the junction-tree variational autoencoder (JT-VAE), a popular model for generative molecular design, and address this issue by leveraging the low dimensional active subspace to capture the uncertainty in the model parameters. Specifically, we approximate the posterior distribution over the active subspace parameters to estimate the epistemic model uncertainty in an extremely high dimensional parameter space. The proposed UQ scheme does not require alteration of the model architecture, making it readily applicable to any pre-trained model. Our experiments demonstrate the efficacy of the AS-based UQ and its potential impact on molecular optimization by exploring the model diversity under epistemic uncertainty.
Multi-Objective Latent Space Optimization of Generative Molecular Design Models
Mar 01, 2022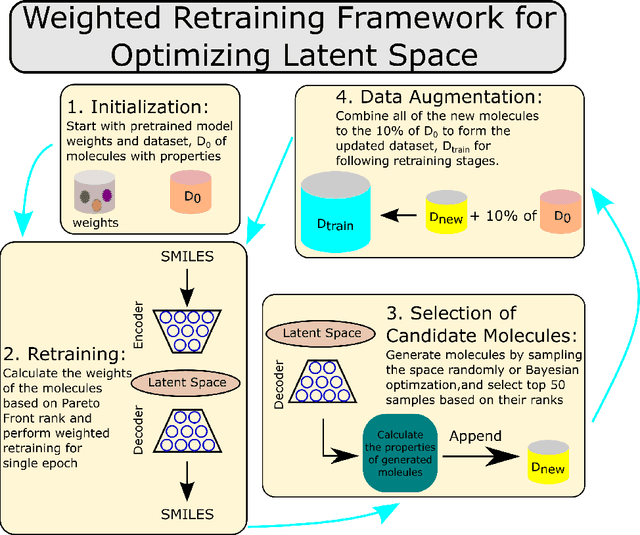
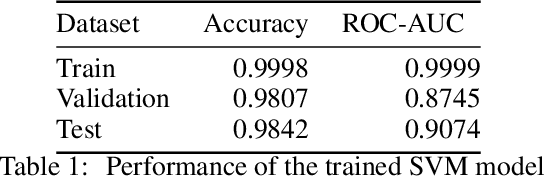
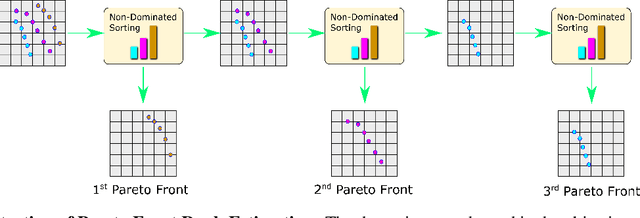
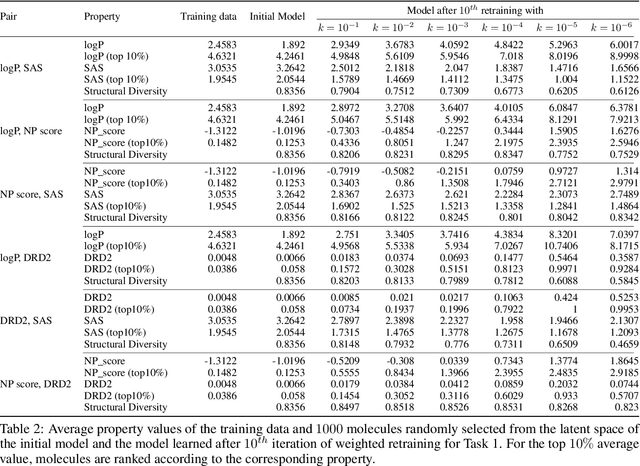
Abstract:Molecular design based on generative models, such as variational autoencoders (VAEs), has become increasingly popular in recent years due to its efficiency for exploring high-dimensional molecular space to identify molecules with desired properties. While the efficacy of the initial model strongly depends on the training data, the sampling efficiency of the model for suggesting novel molecules with enhanced properties can be further enhanced via latent space optimization. In this paper, we propose a multi-objective latent space optimization (LSO) method that can significantly enhance the performance of generative molecular design (GMD). The proposed method adopts an iterative weighted retraining approach, where the respective weights of the molecules in the training data are determined by their Pareto efficiency. We demonstrate that our multi-objective GMD LSO method can significantly improve the performance of GMD for jointly optimizing multiple molecular properties.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge