Brandon Anderson
Integrating Reward Maximization and Population Estimation: Sequential Decision-Making for Internal Revenue Service Audit Selection
Apr 25, 2022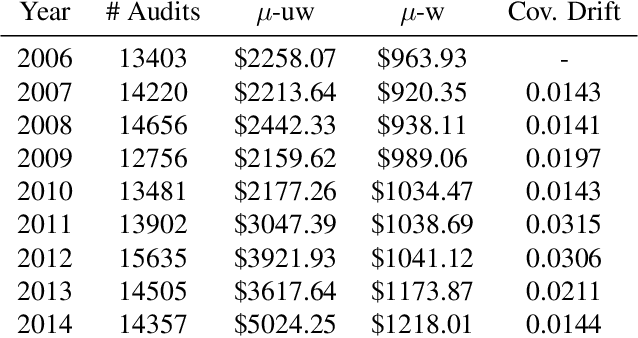
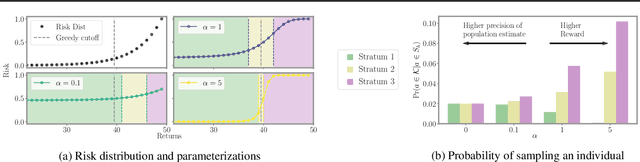
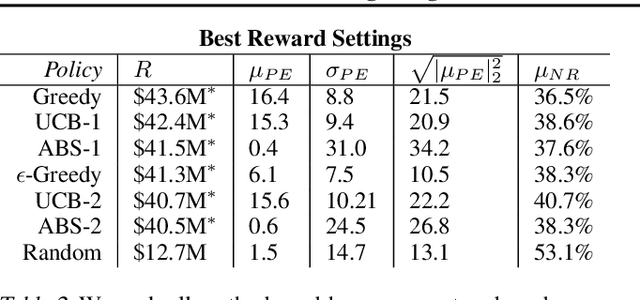
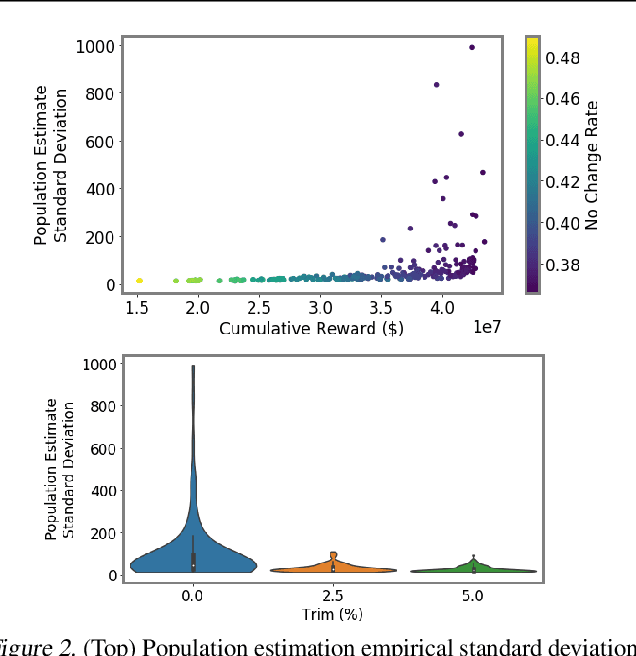
Abstract:We introduce a new setting, optimize-and-estimate structured bandits. Here, a policy must select a batch of arms, each characterized by its own context, that would allow it to both maximize reward and maintain an accurate (ideally unbiased) population estimate of the reward. This setting is inherent to many public and private sector applications and often requires handling delayed feedback, small data, and distribution shifts. We demonstrate its importance on real data from the United States Internal Revenue Service (IRS). The IRS performs yearly audits of the tax base. Two of its most important objectives are to identify suspected misreporting and to estimate the "tax gap" - the global difference between the amount paid and true amount owed. We cast these two processes as a unified optimize-and-estimate structured bandit. We provide a novel mechanism for unbiased population estimation that achieves rewards comparable to baseline approaches. This approach has the potential to improve audit efficacy, while maintaining policy-relevant estimates of the tax gap. This has important social consequences given that the current tax gap is estimated at nearly half a trillion dollars. We suggest that this problem setting is fertile ground for further research and we highlight its interesting challenges.
Mapping industrial poultry operations at scale with deep learning and aerial imagery
Dec 21, 2021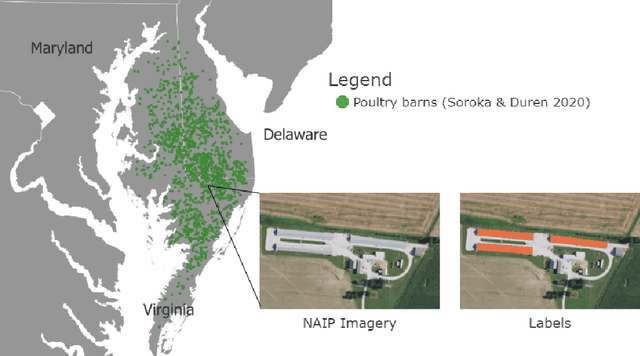
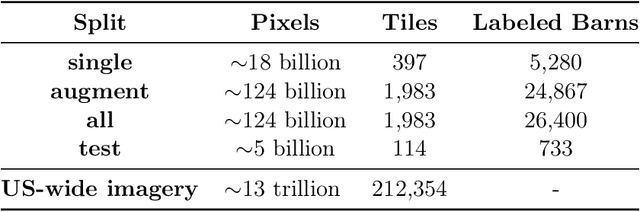
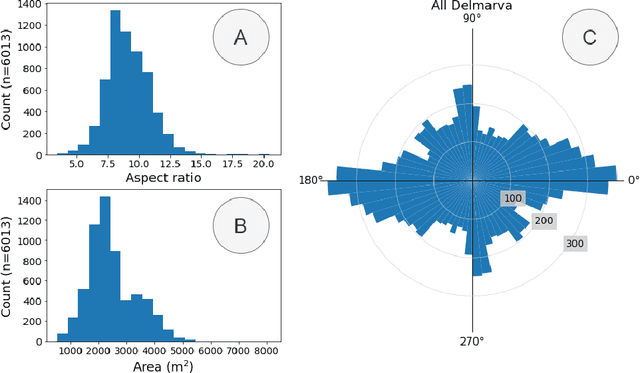

Abstract:Concentrated Animal Feeding Operations (CAFOs) pose serious risks to air, water, and public health, but have proven to be challenging to regulate. The U.S. Government Accountability Office notes that a basic challenge is the lack of comprehensive location information on CAFOs. We use the USDA's National Agricultural Imagery Program (NAIP) 1m/pixel aerial imagery to detect poultry CAFOs across the continental United States. We train convolutional neural network (CNN) models to identify individual poultry barns and apply the best performing model to over 42 TB of imagery to create the first national, open-source dataset of poultry CAFOs. We validate the model predictions against held-out validation set on poultry CAFO facility locations from 10 hand-labeled counties in California and demonstrate that this approach has significant potential to fill gaps in environmental monitoring.
Enhancing Environmental Enforcement with Near Real-Time Monitoring: Likelihood-Based Detection of Structural Expansion of Intensive Livestock Farms
May 29, 2021
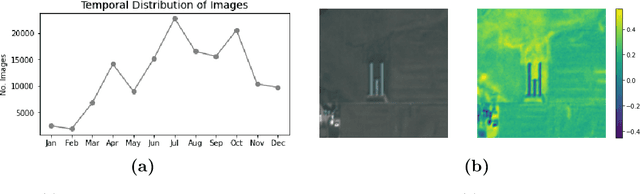
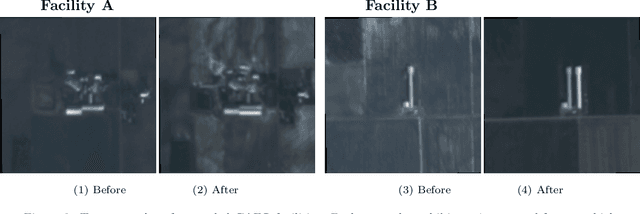
Abstract:Environmental enforcement has historically relied on physical, resource-intensive, and infrequent inspections. Advances in remote sensing and computer vision have the potential to augment compliance monitoring, by providing early warning signals of permit violations. We demonstrate a process for rapid identification of significant structural expansion using satellite imagery and focusing on Concentrated Animal Feeding Operations (CAFOs) as a test case. Unpermitted expansion has been a particular challenge with CAFOs, which pose significant health and environmental risks. Using a new hand-labeled dataset of 175,736 images of 1,513 CAFOs, we combine state-of-the-art building segmentation with a likelihood-based change-point detection model to provide a robust signal of building expansion (AUC = 0.80). A major advantage of this approach is that it is able to work with high-cadence (daily to weekly), but lower resolution (3m/pixel), satellite imagery. It is also highly generalizable and thus provides a near real-time monitoring tool to prioritize enforcement resources to other settings where unpermitted construction poses environmental risk, e.g. zoning, habitat modification, or wetland protection.
Temporal Cluster Matching for Change Detection of Structures from Satellite Imagery
Mar 17, 2021



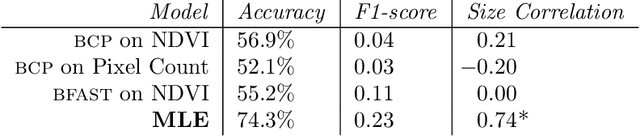
Abstract:We propose a general model, Temporal Cluster Matching (TCM), for detecting building changes in time series of remotely sensed imagery when footprint labels are only available for a single point in time. The intuition behind the model is that the relationship between spectral values inside and outside of building's footprint will change when a building is constructed (or demolished). For instance, in rural settings, the pre-construction area may look similar to the surrounding environment until the building is constructed. Similarly, in urban settings, the pre-construction areas will look different from the surrounding environment until construction. We further propose a heuristic method for selecting the parameters of our model which allows it to be applied in novel settings without requiring data labeling efforts (to fit the parameters). We apply our model over a dataset of poultry barns from 2016/2017 high-resolution aerial imagery in the Delmarva Peninsula and a dataset of solar farms from a 2020 mosaic of Sentinel 2 imagery in India. Our results show that our model performs as well when fit using the proposed heuristic as it does when fit with labeled data, and further, that supervised versions of our model perform the best among all the baselines we test against. Finally, we show that our proposed approach can act as an effective data augmentation strategy -- it enables researchers to augment existing structure footprint labels along the time dimension and thus use imagery from multiple points in time to train deep learning models. We show that this improves the spatial generalization of such models when evaluated on the same change detection task.
ATOM3D: Tasks On Molecules in Three Dimensions
Dec 07, 2020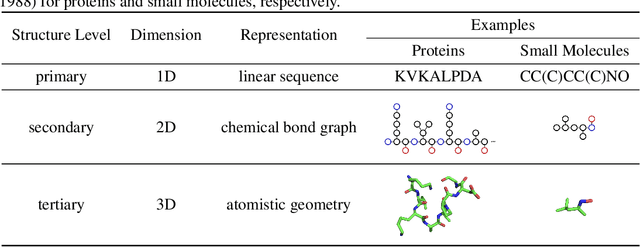
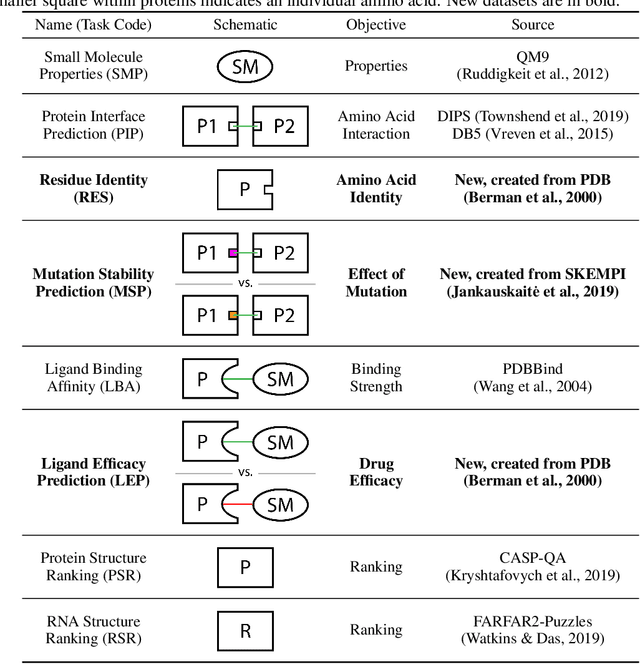
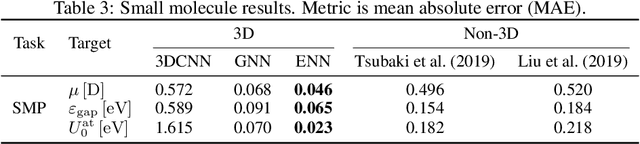
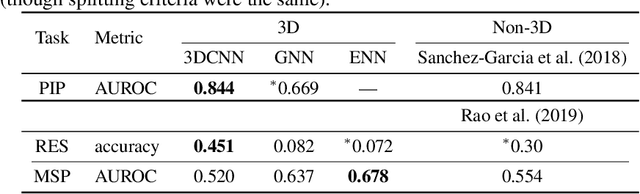
Abstract:Computational methods that operate directly on three-dimensional molecular structure hold large potential to solve important questions in biology and chemistry. In particular deep neural networks have recently gained significant attention. In this work we present ATOM3D, a collection of both novel and existing datasets spanning several key classes of biomolecules, to systematically assess such learning methods. We develop three-dimensional molecular learning networks for each of these tasks, finding that they consistently improve performance relative to one- and two-dimensional methods. The specific choice of architecture proves to be critical for performance, with three-dimensional convolutional networks excelling at tasks involving complex geometries, while graph networks perform well on systems requiring detailed positional information. Furthermore, equivariant networks show significant promise. Our results indicate many molecular problems stand to gain from three-dimensional molecular learning. All code and datasets can be accessed via https://www.atom3d.ai .
A community-powered search of machine learning strategy space to find NMR property prediction models
Aug 13, 2020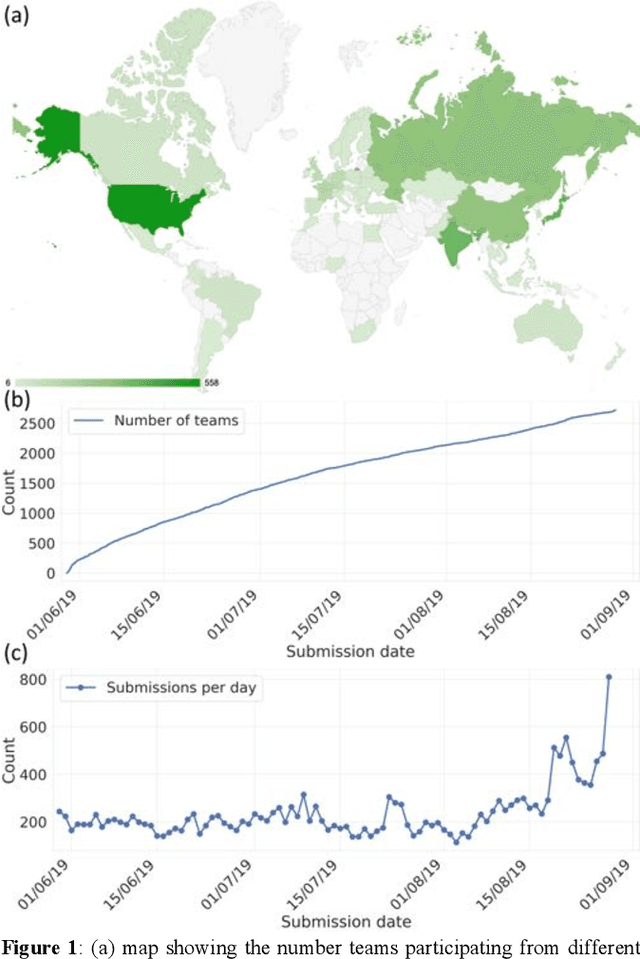
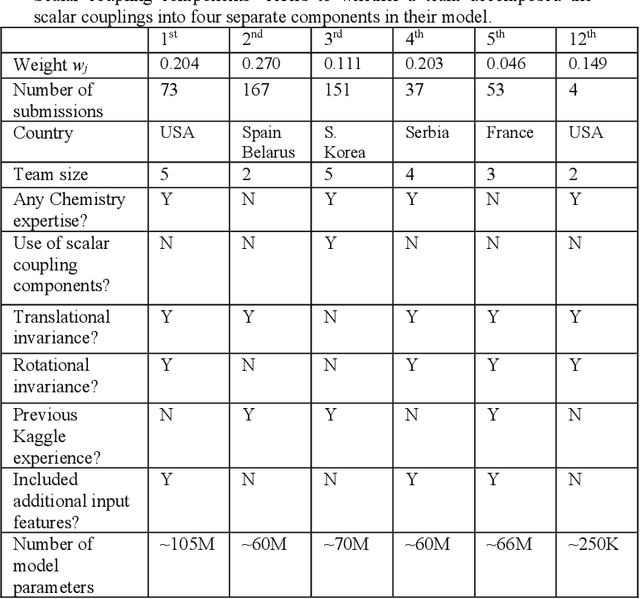
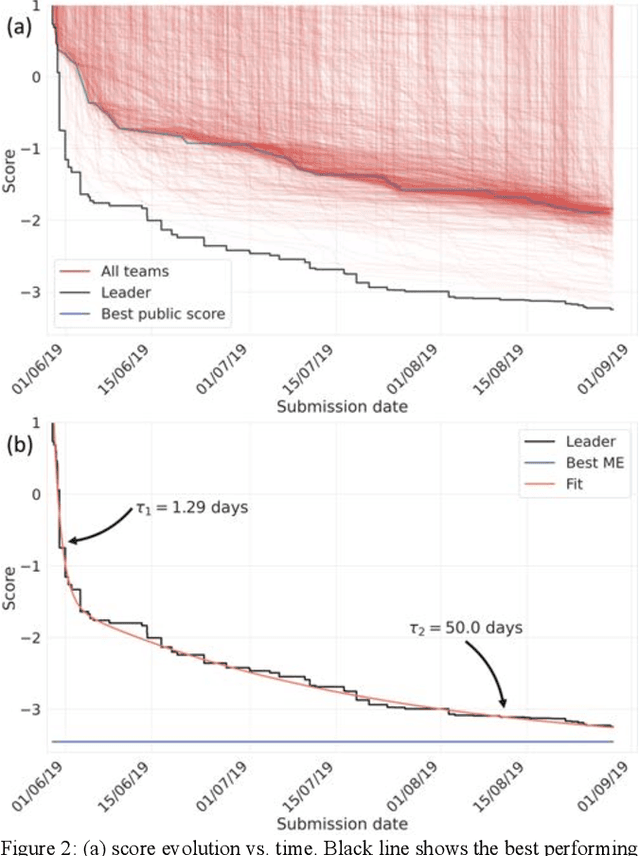
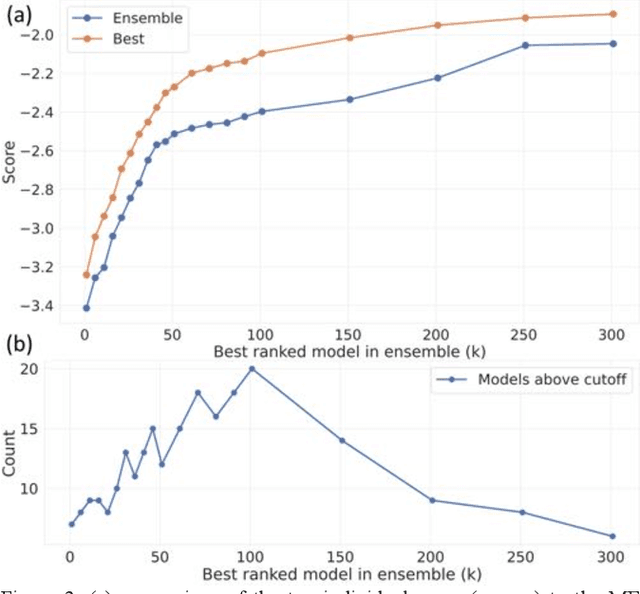
Abstract:The rise of machine learning (ML) has created an explosion in the potential strategies for using data to make scientific predictions. For physical scientists wishing to apply ML strategies to a particular domain, it can be difficult to assess in advance what strategy to adopt within a vast space of possibilities. Here we outline the results of an online community-powered effort to swarm search the space of ML strategies and develop algorithms for predicting atomic-pairwise nuclear magnetic resonance (NMR) properties in molecules. Using an open-source dataset, we worked with Kaggle to design and host a 3-month competition which received 47,800 ML model predictions from 2,700 teams in 84 countries. Within 3 weeks, the Kaggle community produced models with comparable accuracy to our best previously published "in-house" efforts. A meta-ensemble model constructed as a linear combination of the top predictions has a prediction accuracy which exceeds that of any individual model, 7-19x better than our previous state-of-the-art. The results highlight the potential of transformer architectures for predicting quantum mechanical (QM) molecular properties.
Lorentz Group Equivariant Neural Network for Particle Physics
Jun 08, 2020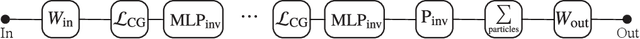
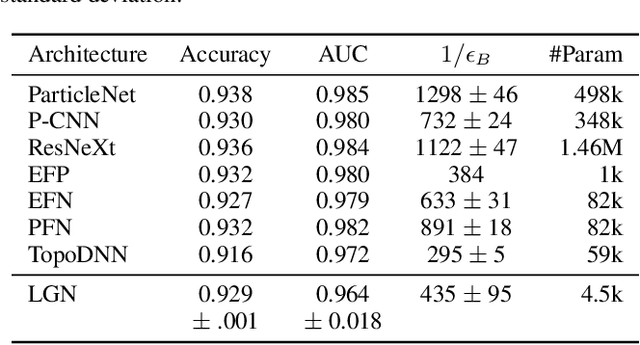
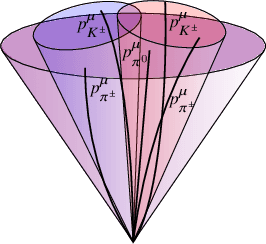
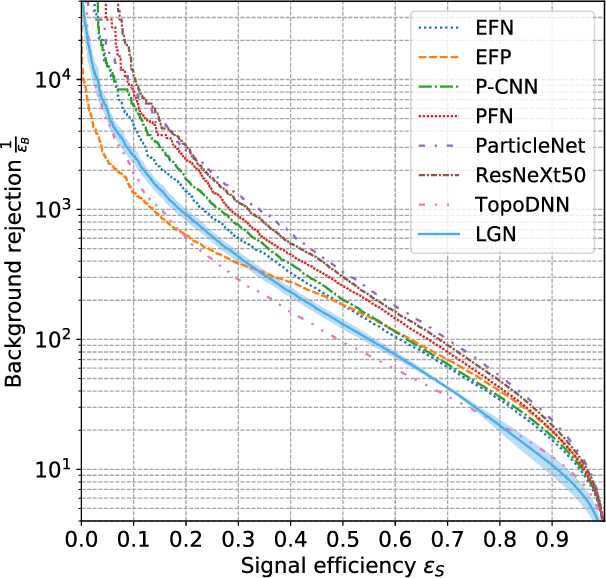
Abstract:We present a neural network architecture that is fully equivariant with respect to transformations under the Lorentz group, a fundamental symmetry of space and time in physics. The architecture is based on the theory of the finite-dimensional representations of the Lorentz group and the equivariant nonlinearity involves the tensor product. For classification tasks in particle physics, we demonstrate that such an equivariant architecture leads to drastically simpler models that have relatively few learnable parameters and are much more physically interpretable than leading approaches that use CNNs and point cloud approaches. The competitive performance of the network is demonstrated on a public classification dataset [27] for tagging top quark decays given energy-momenta of jet constituents produced in proton-proton collisions.
Cormorant: Covariant Molecular Neural Networks
Jun 06, 2019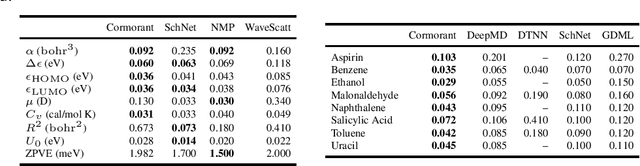
Abstract:We propose Cormorant, a rotationally covariant neural network architecture for learning the behavior and properties of complex many-body physical systems. We apply these networks to molecular systems with two goals: learning atomic potential energy surfaces for use in Molecular Dynamics simulations, and learning ground state properties of molecules calculated by Density Functional Theory. Some of the key features of our network are that (a) each neuron explicitly corresponds to a subset of atoms; (b) the activation of each neuron is covariant to rotations, ensuring that overall the network is fully rotationally invariant. Furthermore, the non-linearity in our network is based upon tensor products and the Clebsch-Gordan decomposition, allowing the network to operate entirely in Fourier space. Cormorant significantly outperforms competing algorithms in learning molecular Potential Energy Surfaces from conformational geometries in the MD-17 dataset, and is competitive with other methods at learning geometric, energetic, electronic, and thermodynamic properties of molecules on the GDB-9 dataset.
Covariant Compositional Networks For Learning Graphs
Jan 07, 2018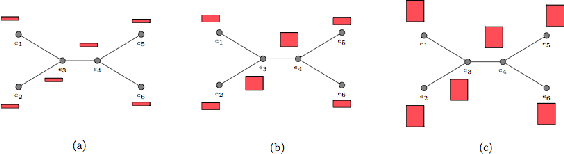
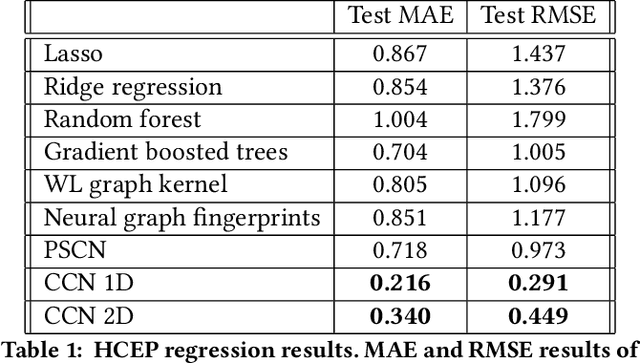
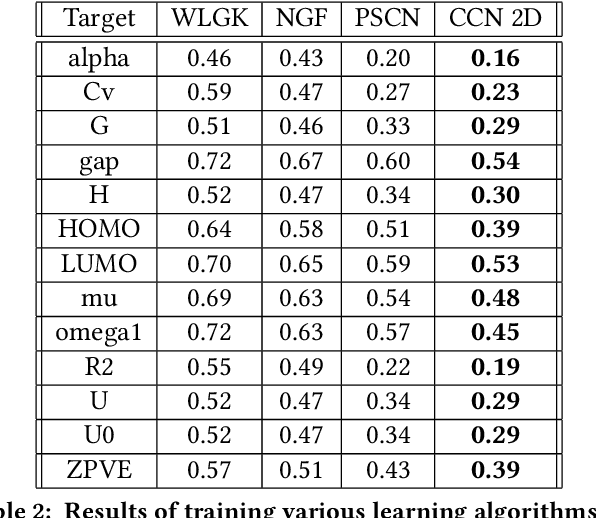
Abstract:Most existing neural networks for learning graphs address permutation invariance by conceiving of the network as a message passing scheme, where each node sums the feature vectors coming from its neighbors. We argue that this imposes a limitation on their representation power, and instead propose a new general architecture for representing objects consisting of a hierarchy of parts, which we call Covariant Compositional Networks (CCNs). Here, covariance means that the activation of each neuron must transform in a specific way under permutations, similarly to steerability in CNNs. We achieve covariance by making each activation transform according to a tensor representation of the permutation group, and derive the corresponding tensor aggregation rules that each neuron must implement. Experiments show that CCNs can outperform competing methods on standard graph learning benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge