Sidhika Balachandar
Urban Incident Prediction with Graph Neural Networks: Integrating Government Ratings and Crowdsourced Reports
Jun 10, 2025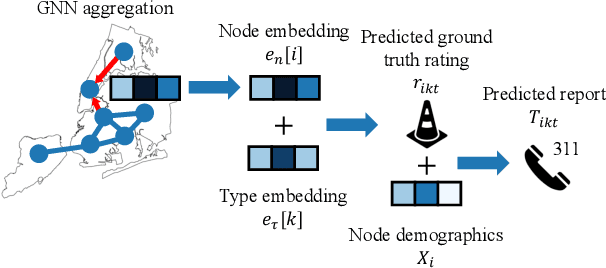
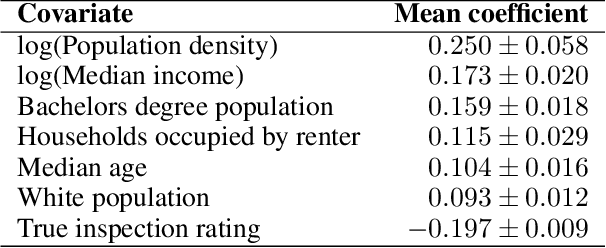
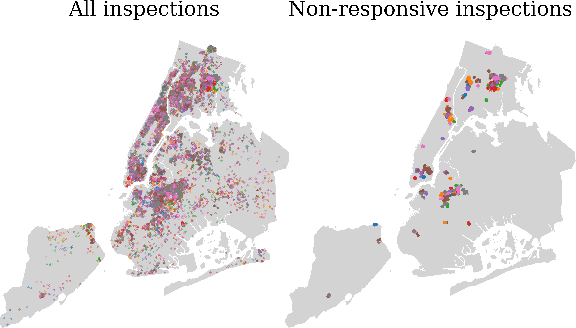
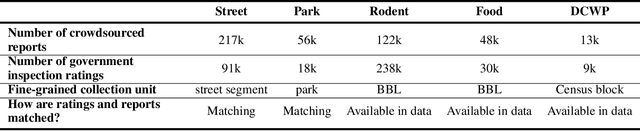
Abstract:Graph neural networks (GNNs) are widely used in urban spatiotemporal forecasting, such as predicting infrastructure problems. In this setting, government officials wish to know in which neighborhoods incidents like potholes or rodent issues occur. The true state of incidents (e.g., street conditions) for each neighborhood is observed via government inspection ratings. However, these ratings are only conducted for a sparse set of neighborhoods and incident types. We also observe the state of incidents via crowdsourced reports, which are more densely observed but may be biased due to heterogeneous reporting behavior. First, for such settings, we propose a multiview, multioutput GNN-based model that uses both unbiased rating data and biased reporting data to predict the true latent state of incidents. Second, we investigate a case study of New York City urban incidents and collect, standardize, and make publicly available a dataset of 9,615,863 crowdsourced reports and 1,041,415 government inspection ratings over 3 years and across 139 types of incidents. Finally, we show on both real and semi-synthetic data that our model can better predict the latent state compared to models that use only reporting data or models that use only rating data, especially when rating data is sparse and reports are predictive of ratings. We also quantify demographic biases in crowdsourced reporting, e.g., higher-income neighborhoods report problems at higher rates. Our analysis showcases a widely applicable approach for latent state prediction using heterogeneous, sparse, and biased data.
Domain constraints improve risk prediction when outcome data is missing
Dec 06, 2023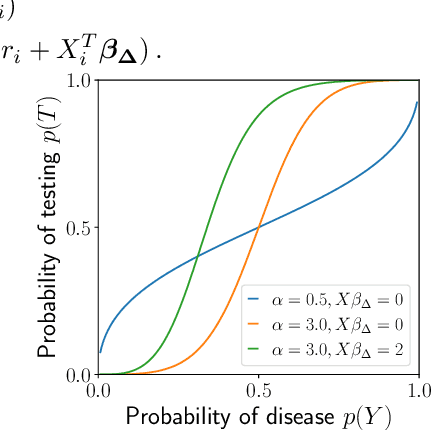

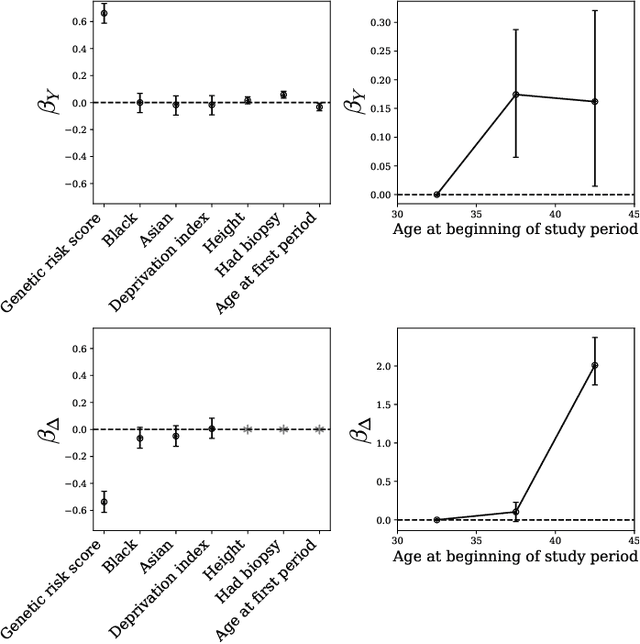
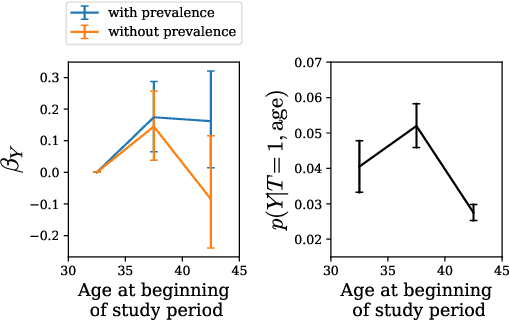
Abstract:Machine learning models are often trained to predict the outcome resulting from a human decision. For example, if a doctor decides to test a patient for disease, will the patient test positive? A challenge is that the human decision censors the outcome data: we only observe test outcomes for patients doctors historically tested. Untested patients, for whom outcomes are unobserved, may differ from tested patients along observed and unobserved dimensions. We propose a Bayesian model class which captures this setting. The purpose of the model is to accurately estimate risk for both tested and untested patients. Estimating this model is challenging due to the wide range of possibilities for untested patients. To address this, we propose two domain constraints which are plausible in health settings: a prevalence constraint, where the overall disease prevalence is known, and an expertise constraint, where the human decision-maker deviates from purely risk-based decision-making only along a constrained feature set. We show theoretically and on synthetic data that domain constraints improve parameter inference. We apply our model to a case study of cancer risk prediction, showing that the model's inferred risk predicts cancer diagnoses, its inferred testing policy captures known public health policies, and it can identify suboptimalities in test allocation. Though our case study is in healthcare, our analysis reveals a general class of domain constraints which can improve model estimation in many settings.
Large language models shape and are shaped by society: A survey of arXiv publication patterns
Jul 20, 2023Abstract:There has been a steep recent increase in the number of large language model (LLM) papers, producing a dramatic shift in the scientific landscape which remains largely undocumented through bibliometric analysis. Here, we analyze 388K papers posted on the CS and Stat arXivs, focusing on changes in publication patterns in 2023 vs. 2018-2022. We analyze how the proportion of LLM papers is increasing; the LLM-related topics receiving the most attention; the authors writing LLM papers; how authors' research topics correlate with their backgrounds; the factors distinguishing highly cited LLM papers; and the patterns of international collaboration. We show that LLM research increasingly focuses on societal impacts: there has been an 18x increase in the proportion of LLM-related papers on the Computers and Society sub-arXiv, and authors newly publishing on LLMs are more likely to focus on applications and societal impacts than more experienced authors. LLM research is also shaped by social dynamics: we document gender and academic/industry disparities in the topics LLM authors focus on, and a US/China schism in the collaboration network. Overall, our analysis documents the profound ways in which LLM research both shapes and is shaped by society, attesting to the necessity of sociotechnical lenses.
Breaking the Symmetry: Resolving Symmetry Ambiguities in Equivariant Neural Networks
Oct 29, 2022



Abstract:Equivariant networks have been adopted in many 3-D learning areas. Here we identify a fundamental limitation of these networks: their ambiguity to symmetries. Equivariant networks cannot complete symmetry-dependent tasks like segmenting a left-right symmetric object into its left and right sides. We tackle this problem by adding components that resolve symmetry ambiguities while preserving rotational equivariance. We present OAVNN: Orientation Aware Vector Neuron Network, an extension of the Vector Neuron Network. OAVNN is a rotation equivariant network that is robust to planar symmetric inputs. Our network consists of three key components. 1) We introduce an algorithm to calculate symmetry detecting features. 2) We create a symmetry-sensitive orientation aware linear layer. 3) We construct an attention mechanism that relates directional information across points. We evaluate the network using left-right segmentation and find that the network quickly obtains accurate segmentations. We hope this work motivates investigations on the expressivity of equivariant networks on symmetric objects.
ATOM3D: Tasks On Molecules in Three Dimensions
Dec 07, 2020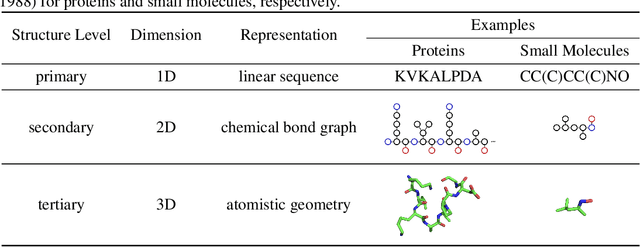
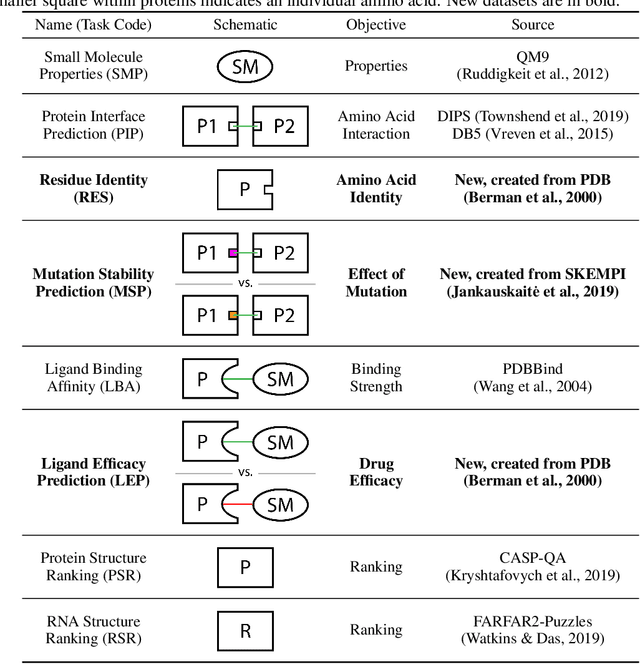
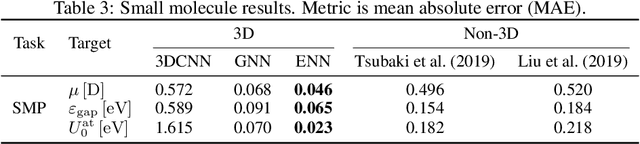
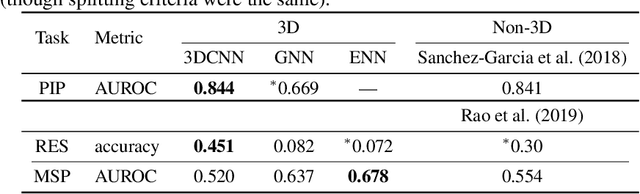
Abstract:Computational methods that operate directly on three-dimensional molecular structure hold large potential to solve important questions in biology and chemistry. In particular deep neural networks have recently gained significant attention. In this work we present ATOM3D, a collection of both novel and existing datasets spanning several key classes of biomolecules, to systematically assess such learning methods. We develop three-dimensional molecular learning networks for each of these tasks, finding that they consistently improve performance relative to one- and two-dimensional methods. The specific choice of architecture proves to be critical for performance, with three-dimensional convolutional networks excelling at tasks involving complex geometries, while graph networks perform well on systems requiring detailed positional information. Furthermore, equivariant networks show significant promise. Our results indicate many molecular problems stand to gain from three-dimensional molecular learning. All code and datasets can be accessed via https://www.atom3d.ai .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge