Zhi Huang
iSight: Towards expert-AI co-assessment for improved immunohistochemistry staining interpretation
Feb 03, 2026Abstract:Immunohistochemistry (IHC) provides information on protein expression in tissue sections and is commonly used to support pathology diagnosis and disease triage. While AI models for H\&E-stained slides show promise, their applicability to IHC is limited due to domain-specific variations. Here we introduce HPA10M, a dataset that contains 10,495,672 IHC images from the Human Protein Atlas with comprehensive metadata included, and encompasses 45 normal tissue types and 20 major cancer types. Based on HPA10M, we trained iSight, a multi-task learning framework for automated IHC staining assessment. iSight combines visual features from whole-slide images with tissue metadata through a token-level attention mechanism, simultaneously predicting staining intensity, location, quantity, tissue type, and malignancy status. On held-out data, iSight achieved 85.5\% accuracy for location, 76.6\% for intensity, and 75.7\% for quantity, outperforming fine-tuned foundation models (PLIP, CONCH) by 2.5--10.2\%. In addition, iSight demonstrates well-calibrated predictions with expected calibration errors of 0.0150-0.0408. Furthermore, in a user study with eight pathologists evaluating 200 images from two datasets, iSight outperformed initial pathologist assessments on the held-out HPA dataset (79\% vs 68\% for location, 70\% vs 57\% for intensity, 68\% vs 52\% for quantity). Inter-pathologist agreement also improved after AI assistance in both held-out HPA (Cohen's $κ$ increased from 0.63 to 0.70) and Stanford TMAD datasets (from 0.74 to 0.76), suggesting expert--AI co-assessment can improve IHC interpretation. This work establishes a foundation for AI systems that can improve IHC diagnostic accuracy and highlights the potential for integrating iSight into clinical workflows to enhance the consistency and reliability of IHC assessment.
VISTA-PATH: An interactive foundation model for pathology image segmentation and quantitative analysis in computational pathology
Jan 23, 2026Abstract:Accurate semantic segmentation for histopathology image is crucial for quantitative tissue analysis and downstream clinical modeling. Recent segmentation foundation models have improved generalization through large-scale pretraining, yet remain poorly aligned with pathology because they treat segmentation as a static visual prediction task. Here we present VISTA-PATH, an interactive, class-aware pathology segmentation foundation model designed to resolve heterogeneous structures, incorporate expert feedback, and produce pixel-level segmentation that are directly meaningful for clinical interpretation. VISTA-PATH jointly conditions segmentation on visual context, semantic tissue descriptions, and optional expert-provided spatial prompts, enabling precise multi-class segmentation across heterogeneous pathology images. To support this paradigm, we curate VISTA-PATH Data, a large-scale pathology segmentation corpus comprising over 1.6 million image-mask-text triplets spanning 9 organs and 93 tissue classes. Across extensive held-out and external benchmarks, VISTA-PATH consistently outperforms existing segmentation foundation models. Importantly, VISTA-PATH supports dynamic human-in-the-loop refinement by propagating sparse, patch-level bounding-box annotation feedback into whole-slide segmentation. Finally, we show that the high-fidelity, class-aware segmentation produced by VISTA-PATH is a preferred model for computational pathology. It improve tissue microenvironment analysis through proposed Tumor Interaction Score (TIS), which exhibits strong and significant associations with patient survival. Together, these results establish VISTA-PATH as a foundation model that elevates pathology image segmentation from a static prediction to an interactive and clinically grounded representation for digital pathology. Source code and demo can be found at https://github.com/zhihuanglab/VISTA-PATH.
Adaptive Multi-Scale Integration Unlocks Robust Cell Annotation in Histopathology Images
Nov 18, 2025Abstract:Identifying cell types and subtypes in routine histopathology is fundamental for understanding disease. Existing tile-based models capture nuclear detail but miss the broader tissue context that influences cell identity. Current human annotations are coarse-grained and uneven across studies, making fine-grained, subtype-level classification difficult. In this study, we build a marker-guided dataset from Xenium spatial transcriptomics with single-cell resolution labels for more than two million cells across eight organs and 16 classes to address the lack of high-quality annotations. Leveraging this data resource, we introduce NuClass, a pathologist workflow inspired framework for cell-wise multi-scale integration of nuclear morphology and microenvironmental context. It combines Path local, which focuses on nuclear morphology from 224x224 pixel crops, and Path global, which models the surrounding 1024x1024 pixel neighborhood, through a learnable gating module that balances local and global information. An uncertainty-guided objective directs the global path to prioritize regions where the local path is uncertain, and we provide calibrated confidence estimates and Grad-CAM maps for interpretability. Evaluated on three fully held-out cohorts, NuClass achieves up to 96 percent F1 for its best-performing class, outperforming strong baselines. Our results demonstrate that multi-scale, uncertainty-aware fusion can bridge the gap between slide-level pathological foundation models and reliable, cell-level phenotype prediction.
Pathology-CoT: Learning Visual Chain-of-Thought Agent from Expert Whole Slide Image Diagnosis Behavior
Oct 06, 2025Abstract:Diagnosing a whole-slide image is an interactive, multi-stage process involving changes in magnification and movement between fields. Although recent pathology foundation models are strong, practical agentic systems that decide what field to examine next, adjust magnification, and deliver explainable diagnoses are still lacking. The blocker is data: scalable, clinically aligned supervision of expert viewing behavior that is tacit and experience-based, not written in textbooks or online, and therefore absent from large language model training. We introduce the AI Session Recorder, which works with standard WSI viewers to unobtrusively record routine navigation and convert the viewer logs into standardized behavioral commands (inspect or peek at discrete magnifications) and bounding boxes. A lightweight human-in-the-loop review turns AI-drafted rationales into the Pathology-CoT dataset, a form of paired "where to look" and "why it matters" supervision produced at roughly six times lower labeling time. Using this behavioral data, we build Pathologist-o3, a two-stage agent that first proposes regions of interest and then performs behavior-guided reasoning. On gastrointestinal lymph-node metastasis detection, it achieved 84.5% precision, 100.0% recall, and 75.4% accuracy, exceeding the state-of-the-art OpenAI o3 model and generalizing across backbones. To our knowledge, this constitutes one of the first behavior-grounded agentic systems in pathology. Turning everyday viewer logs into scalable, expert-validated supervision, our framework makes agentic pathology practical and establishes a path to human-aligned, upgradeable clinical AI.
Can Textual Gradient Work in Federated Learning?
Feb 27, 2025Abstract:Recent studies highlight the promise of LLM-based prompt optimization, especially with TextGrad, which automates differentiation'' via texts and backpropagates textual feedback. This approach facilitates training in various real-world applications that do not support numerical gradient propagation or loss calculation. In this paper, we systematically explore the potential and challenges of incorporating textual gradient into Federated Learning (FL). Our contributions are fourfold. Firstly, we introduce a novel FL paradigm, Federated Textual Gradient (FedTextGrad), that allows clients to upload locally optimized prompts derived from textual gradients, while the server aggregates the received prompts. Unlike traditional FL frameworks, which are designed for numerical aggregation, FedTextGrad is specifically tailored for handling textual data, expanding the applicability of FL to a broader range of problems that lack well-defined numerical loss functions. Secondly, building on this design, we conduct extensive experiments to explore the feasibility of FedTextGrad. Our findings highlight the importance of properly tuning key factors (e.g., local steps) in FL training. Thirdly, we highlight a major challenge in FedTextGrad aggregation: retaining essential information from distributed prompt updates. Last but not least, in response to this issue, we improve the vanilla variant of FedTextGrad by providing actionable guidance to the LLM when summarizing client prompts by leveraging the Uniform Information Density principle. Through this principled study, we enable the adoption of textual gradients in FL for optimizing LLMs, identify important issues, and pinpoint future directions, thereby opening up a new research area that warrants further investigation.
TextGrad: Automatic "Differentiation" via Text
Jun 11, 2024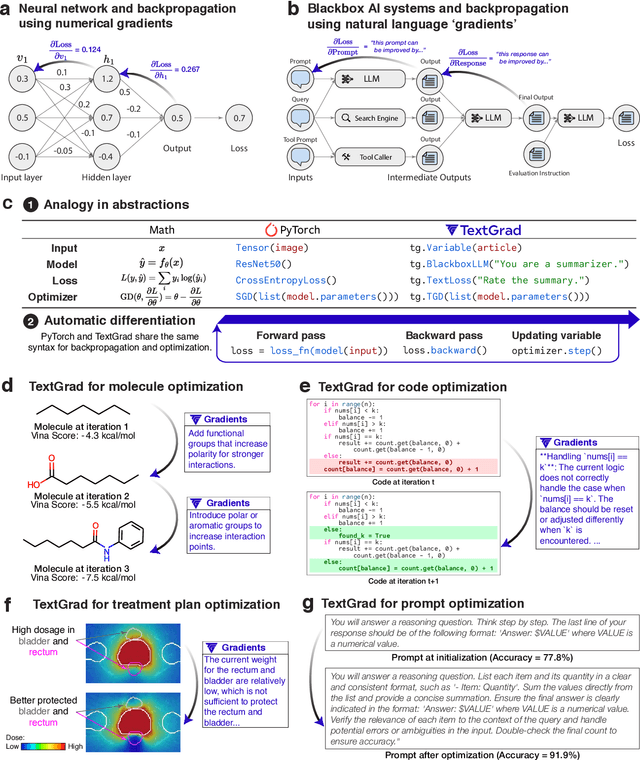

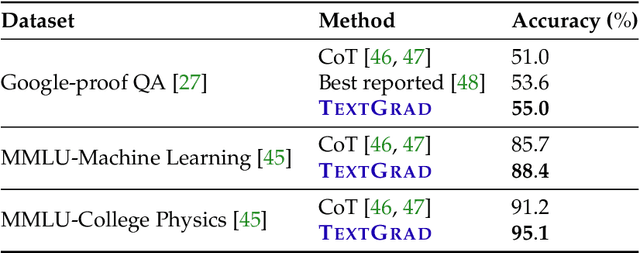
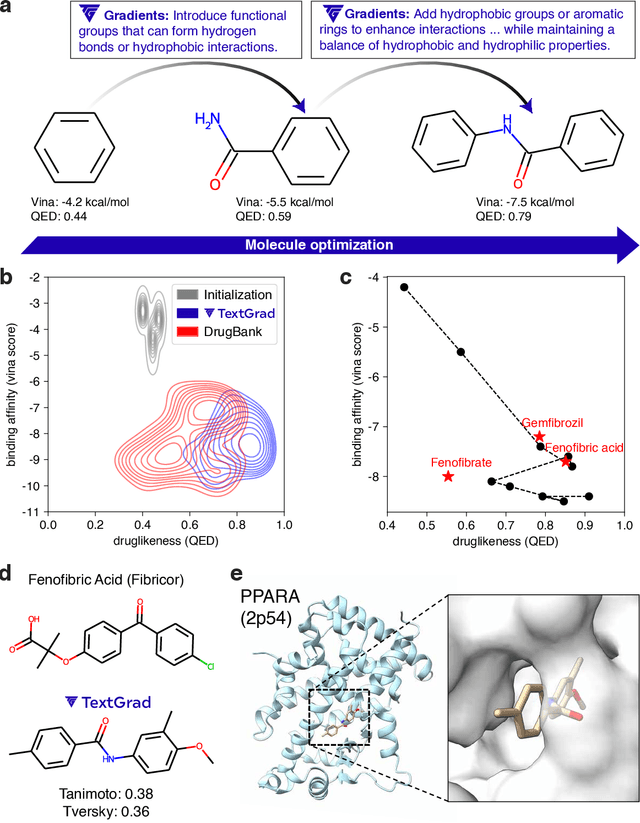
Abstract:AI is undergoing a paradigm shift, with breakthroughs achieved by systems orchestrating multiple large language models (LLMs) and other complex components. As a result, developing principled and automated optimization methods for compound AI systems is one of the most important new challenges. Neural networks faced a similar challenge in its early days until backpropagation and automatic differentiation transformed the field by making optimization turn-key. Inspired by this, we introduce TextGrad, a powerful framework performing automatic ``differentiation'' via text. TextGrad backpropagates textual feedback provided by LLMs to improve individual components of a compound AI system. In our framework, LLMs provide rich, general, natural language suggestions to optimize variables in computation graphs, ranging from code snippets to molecular structures. TextGrad follows PyTorch's syntax and abstraction and is flexible and easy-to-use. It works out-of-the-box for a variety of tasks, where the users only provide the objective function without tuning components or prompts of the framework. We showcase TextGrad's effectiveness and generality across a diverse range of applications, from question answering and molecule optimization to radiotherapy treatment planning. Without modifying the framework, TextGrad improves the zero-shot accuracy of GPT-4o in Google-Proof Question Answering from $51\%$ to $55\%$, yields $20\%$ relative performance gain in optimizing LeetCode-Hard coding problem solutions, improves prompts for reasoning, designs new druglike small molecules with desirable in silico binding, and designs radiation oncology treatment plans with high specificity. TextGrad lays a foundation to accelerate the development of the next-generation of AI systems.
Mapping the Increasing Use of LLMs in Scientific Papers
Apr 01, 2024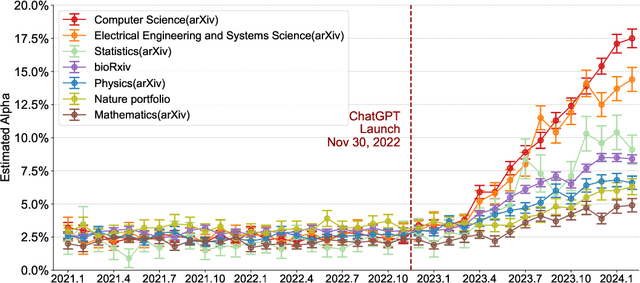
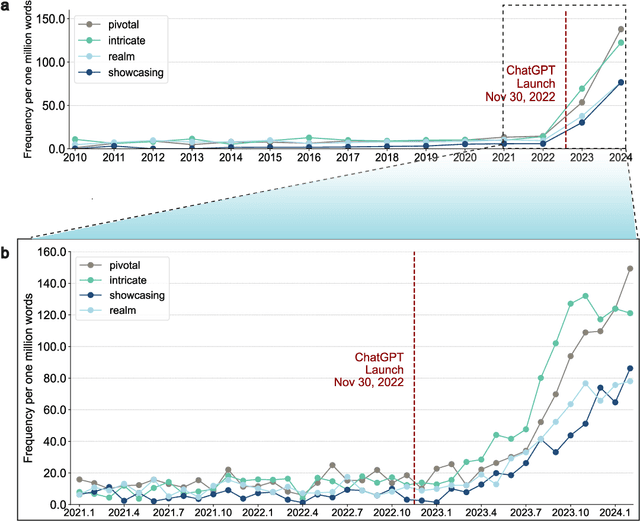
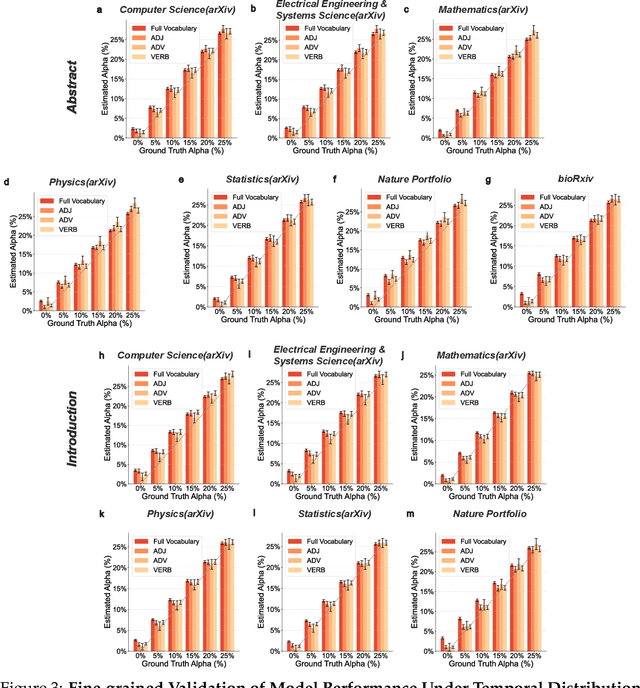
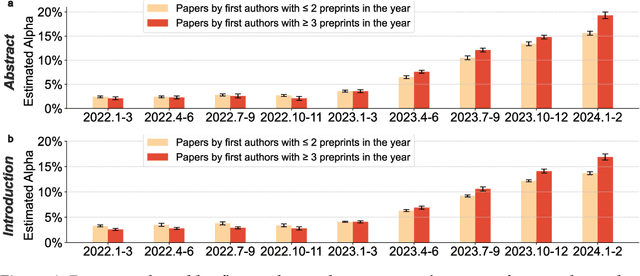
Abstract:Scientific publishing lays the foundation of science by disseminating research findings, fostering collaboration, encouraging reproducibility, and ensuring that scientific knowledge is accessible, verifiable, and built upon over time. Recently, there has been immense speculation about how many people are using large language models (LLMs) like ChatGPT in their academic writing, and to what extent this tool might have an effect on global scientific practices. However, we lack a precise measure of the proportion of academic writing substantially modified or produced by LLMs. To address this gap, we conduct the first systematic, large-scale analysis across 950,965 papers published between January 2020 and February 2024 on the arXiv, bioRxiv, and Nature portfolio journals, using a population-level statistical framework to measure the prevalence of LLM-modified content over time. Our statistical estimation operates on the corpus level and is more robust than inference on individual instances. Our findings reveal a steady increase in LLM usage, with the largest and fastest growth observed in Computer Science papers (up to 17.5%). In comparison, Mathematics papers and the Nature portfolio showed the least LLM modification (up to 6.3%). Moreover, at an aggregate level, our analysis reveals that higher levels of LLM-modification are associated with papers whose first authors post preprints more frequently, papers in more crowded research areas, and papers of shorter lengths. Our findings suggests that LLMs are being broadly used in scientific writings.
Monitoring AI-Modified Content at Scale: A Case Study on the Impact of ChatGPT on AI Conference Peer Reviews
Mar 11, 2024



Abstract:We present an approach for estimating the fraction of text in a large corpus which is likely to be substantially modified or produced by a large language model (LLM). Our maximum likelihood model leverages expert-written and AI-generated reference texts to accurately and efficiently examine real-world LLM-use at the corpus level. We apply this approach to a case study of scientific peer review in AI conferences that took place after the release of ChatGPT: ICLR 2024, NeurIPS 2023, CoRL 2023 and EMNLP 2023. Our results suggest that between 6.5% and 16.9% of text submitted as peer reviews to these conferences could have been substantially modified by LLMs, i.e. beyond spell-checking or minor writing updates. The circumstances in which generated text occurs offer insight into user behavior: the estimated fraction of LLM-generated text is higher in reviews which report lower confidence, were submitted close to the deadline, and from reviewers who are less likely to respond to author rebuttals. We also observe corpus-level trends in generated text which may be too subtle to detect at the individual level, and discuss the implications of such trends on peer review. We call for future interdisciplinary work to examine how LLM use is changing our information and knowledge practices.
A Delay Compensation Framework Based on Eye-Movement for Teleoperated Ground Vehicles
Sep 14, 2023



Abstract:An eye-movement-based predicted trajectory guidance control (ePTGC) is proposed to mitigate the maneuverability degradation of a teleoperated ground vehicle caused by communication delays. Human sensitivity to delays is the main reason for the performance degradation of a ground vehicle teleoperation system. The proposed framework extracts human intention from eye-movement. Then, it combines it with contextual constraints to generate an intention-compliant guidance trajectory, which is then employed to control the vehicle directly. The advantage of this approach is that the teleoperator is removed from the direct control loop by using the generated trajectories to guide vehicle, thus reducing the adverse sensitivity to delay. The delay can be compensated as long as the prediction horizon exceeds the delay. A human-in-loop simulation platform is designed to evaluate the teleoperation performance of the proposed method at different delay levels. The results are analyzed by repeated measures ANOVA, which shows that the proposed method significantly improves maneuverability and cognitive burden at large delay levels (>200 ms). The overall performance is also much better than the PTGC which does not employ the eye-movement feature.
Low-Rank Reorganization via Proportional Hazards Non-negative Matrix Factorization Unveils Survival Associated Gene Clusters
Sep 17, 2020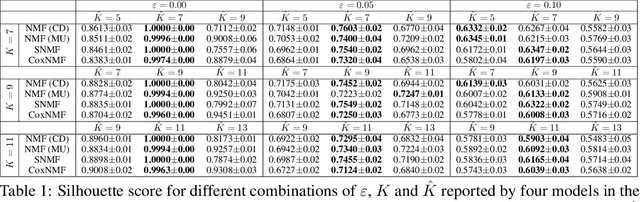
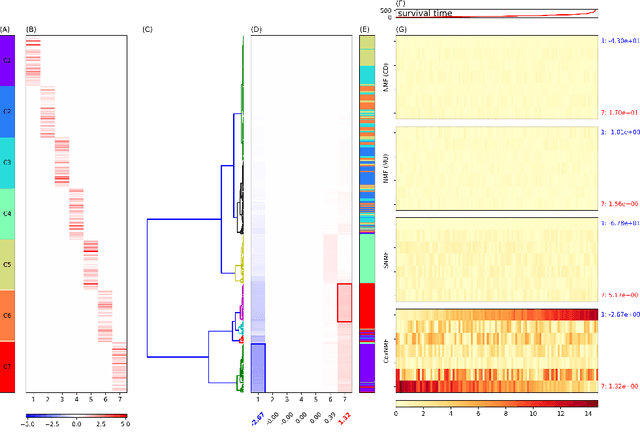
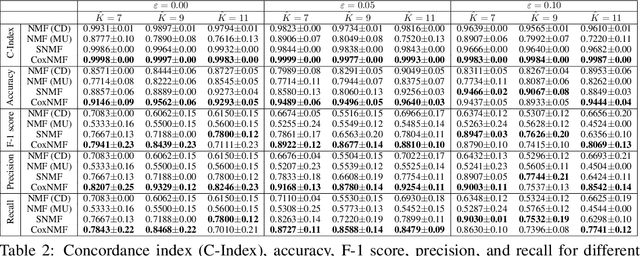
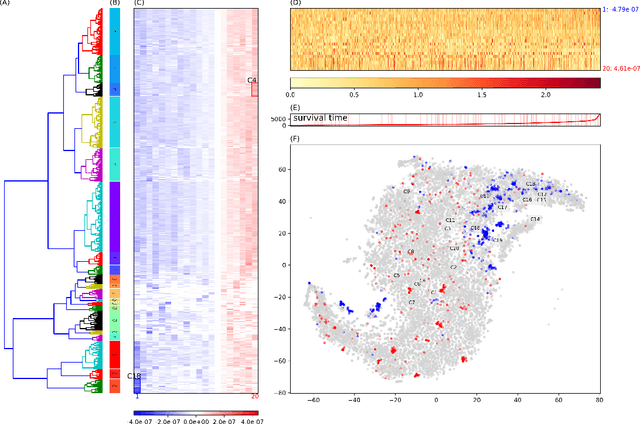
Abstract:One of the central goals in precision health is the understanding and interpretation of high-dimensional biological data to identify genes and markers associated with disease initiation, development, and outcomes. Though significant effort has been committed to harness gene expression data for multiple analyses while accounting for time-to-event modeling by including survival times, many traditional analyses have focused separately on non-negative matrix factorization (NMF) of the gene expression data matrix and survival regression with Cox proportional hazards model. In this work, Cox proportional hazards regression is integrated with NMF by imposing survival constraints. This is accomplished by jointly optimizing the Frobenius norm and partial log likelihood for events such as death or relapse. Simulation results on synthetic data demonstrated the superiority of the proposed method, when compared to other algorithms, in finding survival associated gene clusters. In addition, using human cancer gene expression data, the proposed technique can unravel critical clusters of cancer genes. The discovered gene clusters reflect rich biological implications and can help identify survival-related biomarkers. Towards the goal of precision health and cancer treatments, the proposed algorithm can help understand and interpret high-dimensional heterogeneous genomics data with accurate identification of survival-associated gene clusters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge