Paul Salama
Towards Predicting the Success of Transfer-based Attacks by Quantifying Shared Feature Representations
Dec 06, 2024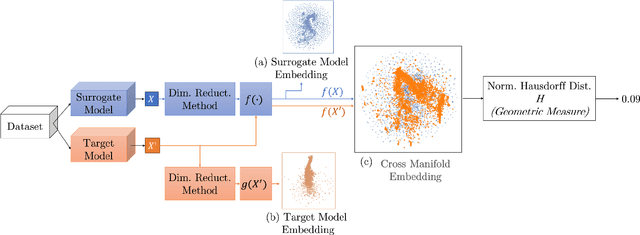
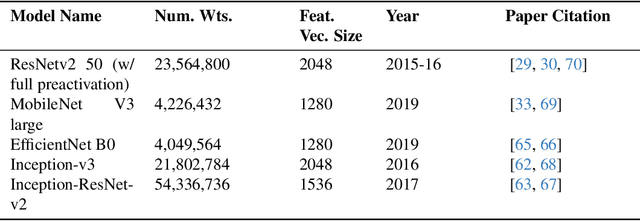
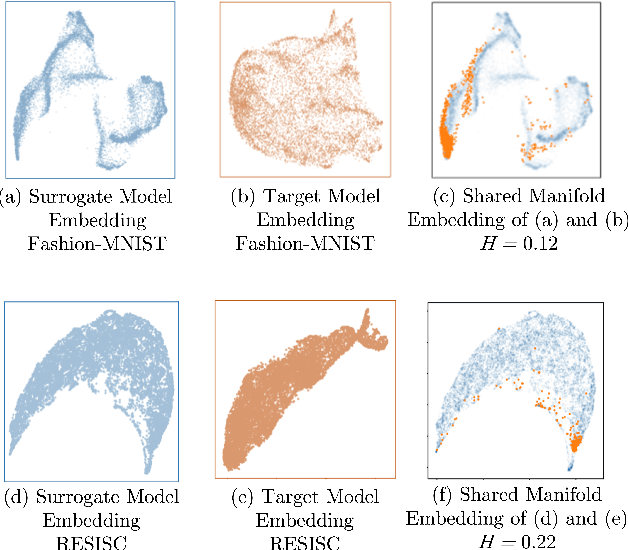

Abstract:Much effort has been made to explain and improve the success of transfer-based attacks (TBA) on black-box computer vision models. This work provides the first attempt at a priori prediction of attack success by identifying the presence of vulnerable features within target models. Recent work by Chen and Liu (2024) proposed the manifold attack model, a unifying framework proposing that successful TBA exist in a common manifold space. Our work experimentally tests the common manifold space hypothesis by a new methodology: first, projecting feature vectors from surrogate and target feature extractors trained on ImageNet onto the same low-dimensional manifold; second, quantifying any observed structure similarities on the manifold; and finally, by relating these observed similarities to the success of the TBA. We find that shared feature representation moderately correlates with increased success of TBA (\r{ho}= 0.56). This method may be used to predict whether an attack will transfer without information of the model weights, training, architecture or details of the attack. The results confirm the presence of shared feature representations between two feature extractors of different sizes and complexities, and demonstrate the utility of datasets from different target domains as test signals for interpreting black-box feature representations.
RCNN-SliceNet: A Slice and Cluster Approach for Nuclei Centroid Detection in Three-Dimensional Fluorescence Microscopy Images
Jun 29, 2021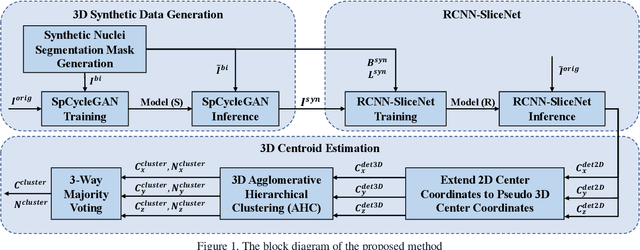

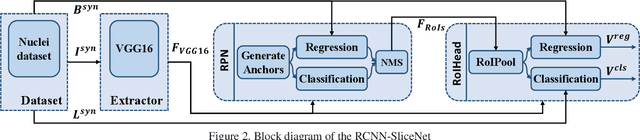

Abstract:Robust and accurate nuclei centroid detection is important for the understanding of biological structures in fluorescence microscopy images. Existing automated nuclei localization methods face three main challenges: (1) Most of object detection methods work only on 2D images and are difficult to extend to 3D volumes; (2) Segmentation-based models can be used on 3D volumes but it is computational expensive for large microscopy volumes and they have difficulty distinguishing different instances of objects; (3) Hand annotated ground truth is limited for 3D microscopy volumes. To address these issues, we present a scalable approach for nuclei centroid detection of 3D microscopy volumes. We describe the RCNN-SliceNet to detect 2D nuclei centroids for each slice of the volume from different directions and 3D agglomerative hierarchical clustering (AHC) is used to estimate the 3D centroids of nuclei in a volume. The model was trained with the synthetic microscopy data generated using Spatially Constrained Cycle-Consistent Adversarial Networks (SpCycleGAN) and tested on different types of real 3D microscopy data. Extensive experimental results demonstrate that our proposed method can accurately count and detect the nuclei centroids in a 3D microscopy volume.
Low-Rank Reorganization via Proportional Hazards Non-negative Matrix Factorization Unveils Survival Associated Gene Clusters
Sep 17, 2020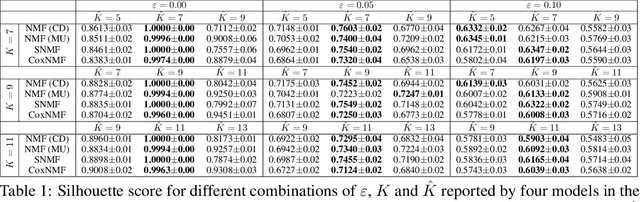
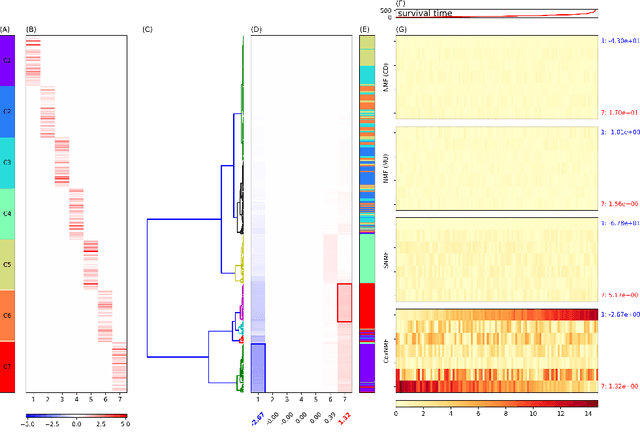
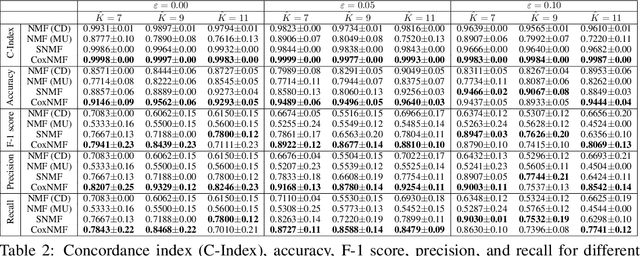
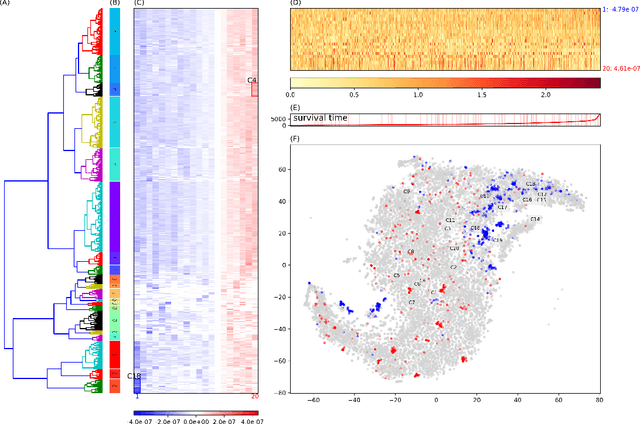
Abstract:One of the central goals in precision health is the understanding and interpretation of high-dimensional biological data to identify genes and markers associated with disease initiation, development, and outcomes. Though significant effort has been committed to harness gene expression data for multiple analyses while accounting for time-to-event modeling by including survival times, many traditional analyses have focused separately on non-negative matrix factorization (NMF) of the gene expression data matrix and survival regression with Cox proportional hazards model. In this work, Cox proportional hazards regression is integrated with NMF by imposing survival constraints. This is accomplished by jointly optimizing the Frobenius norm and partial log likelihood for events such as death or relapse. Simulation results on synthetic data demonstrated the superiority of the proposed method, when compared to other algorithms, in finding survival associated gene clusters. In addition, using human cancer gene expression data, the proposed technique can unravel critical clusters of cancer genes. The discovered gene clusters reflect rich biological implications and can help identify survival-related biomarkers. Towards the goal of precision health and cancer treatments, the proposed algorithm can help understand and interpret high-dimensional heterogeneous genomics data with accurate identification of survival-associated gene clusters.
Center-Extraction-Based Three Dimensional Nuclei Instance Segmentation of Fluorescence Microscopy Images
Sep 13, 2019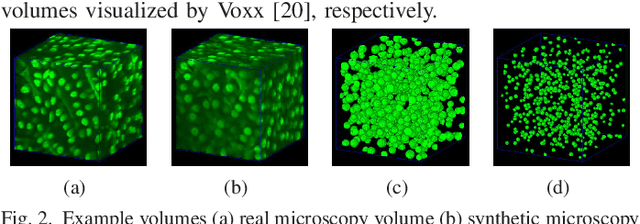
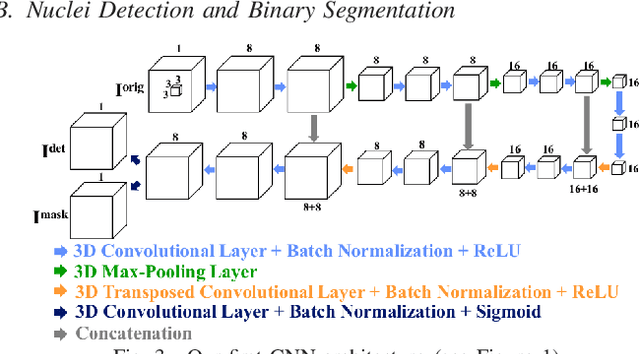
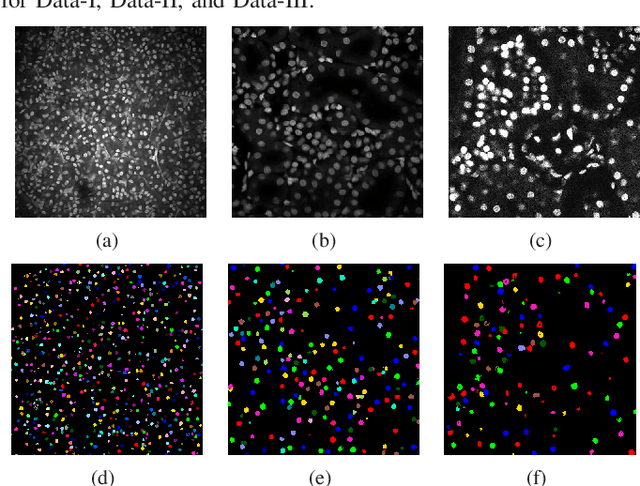
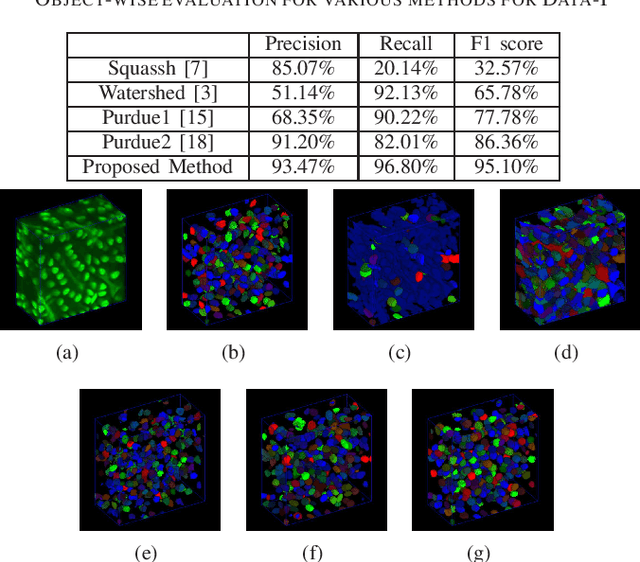
Abstract:Fluorescence microscopy is an essential tool for the analysis of 3D subcellular structures in tissue. An important step in the characterization of tissue involves nuclei segmentation. In this paper, a two-stage method for segmentation of nuclei using convolutional neural networks (CNNs) is described. In particular, since creating labeled volumes manually for training purposes is not practical due to the size and complexity of the 3D data sets, the paper describes a method for generating synthetic microscopy volumes based on a spatially constrained cycle-consistent adversarial network. The proposed method is tested on multiple real microscopy data sets and outperforms other commonly used segmentation techniques.
Three dimensional blind image deconvolution for fluorescence microscopy using generative adversarial networks
Apr 19, 2019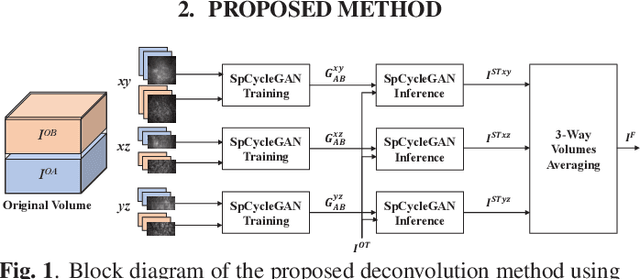

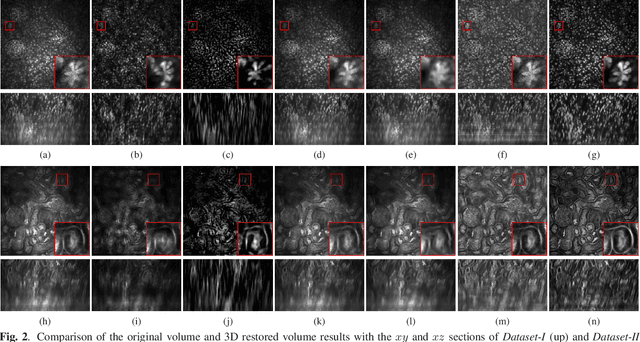
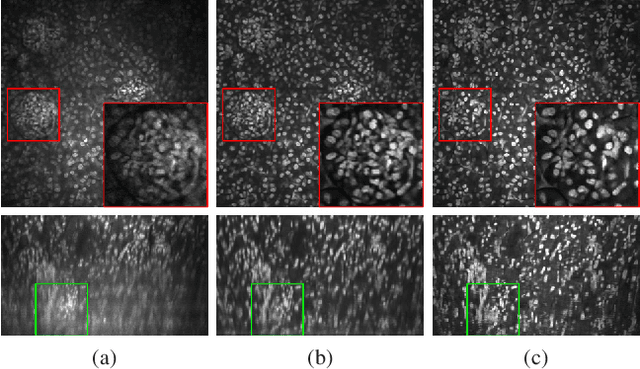
Abstract:Due to image blurring image deconvolution is often used for studying biological structures in fluorescence microscopy. Fluorescence microscopy image volumes inherently suffer from intensity inhomogeneity, blur, and are corrupted by various types of noise which exacerbate image quality at deeper tissue depth. Therefore, quantitative analysis of fluorescence microscopy in deeper tissue still remains a challenge. This paper presents a three dimensional blind image deconvolution method for fluorescence microscopy using 3-way spatially constrained cycle-consistent adversarial networks. The restored volumes of the proposed deconvolution method and other well-known deconvolution methods, denoising methods, and an inhomogeneity correction method are visually and numerically evaluated. Experimental results indicate that the proposed method can restore and improve the quality of blurred and noisy deep depth microscopy image visually and quantitatively.
Three Dimensional Fluorescence Microscopy Image Synthesis and Segmentation
Apr 21, 2018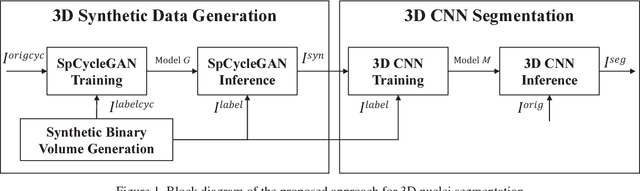
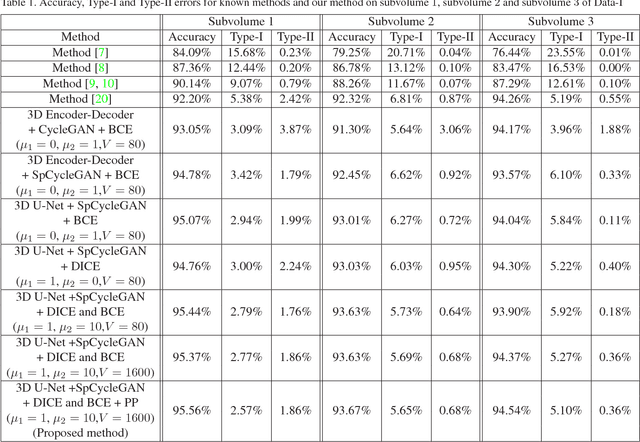
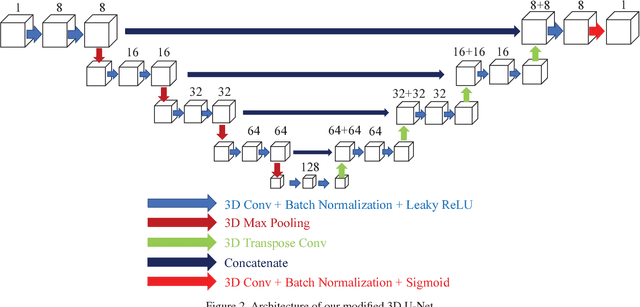
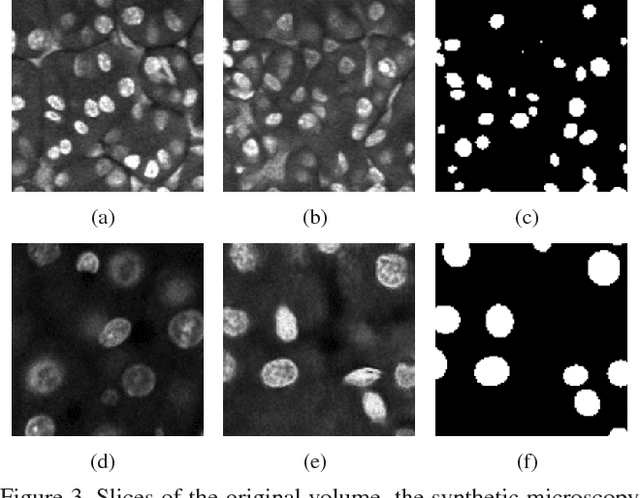
Abstract:Advances in fluorescence microscopy enable acquisition of 3D image volumes with better image quality and deeper penetration into tissue. Segmentation is a required step to characterize and analyze biological structures in the images and recent 3D segmentation using deep learning has achieved promising results. One issue is that deep learning techniques require a large set of groundtruth data which is impractical to annotate manually for large 3D microscopy volumes. This paper describes a 3D deep learning nuclei segmentation method using synthetic 3D volumes for training. A set of synthetic volumes and the corresponding groundtruth are generated using spatially constrained cycle-consistent adversarial networks. Segmentation results demonstrate that our proposed method is capable of segmenting nuclei successfully for various data sets.
Tubule segmentation of fluorescence microscopy images based on convolutional neural networks with inhomogeneity correction
Feb 10, 2018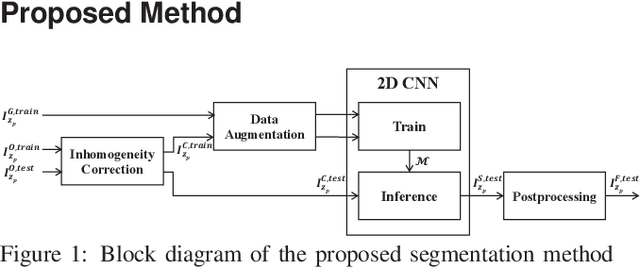
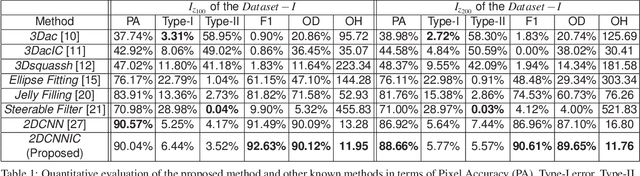
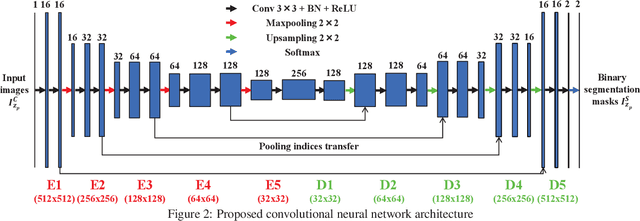

Abstract:Fluorescence microscopy has become a widely used tool for studying various biological structures of in vivo tissue or cells. However, quantitative analysis of these biological structures remains a challenge due to their complexity which is exacerbated by distortions caused by lens aberrations and light scattering. Moreover, manual quantification of such image volumes is an intractable and error-prone process, making the need for automated image analysis methods crucial. This paper describes a segmentation method for tubular structures in fluorescence microscopy images using convolutional neural networks with data augmentation and inhomogeneity correction. The segmentation results of the proposed method are visually and numerically compared with other microscopy segmentation methods. Experimental results indicate that the proposed method has better performance with correctly segmenting and identifying multiple tubular structures compared to other methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge