Chichen Fu
Center-Extraction-Based Three Dimensional Nuclei Instance Segmentation of Fluorescence Microscopy Images
Sep 13, 2019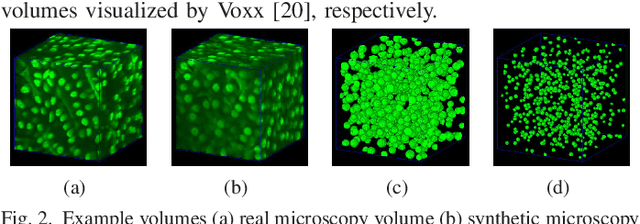
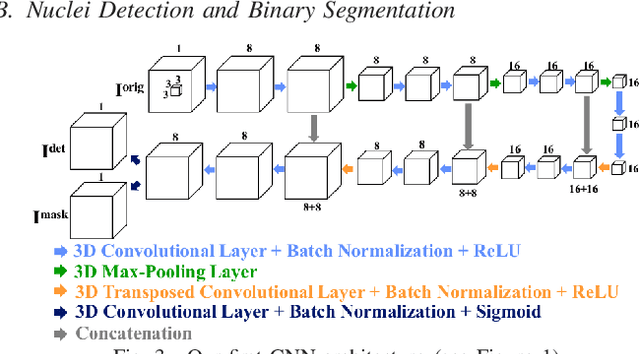
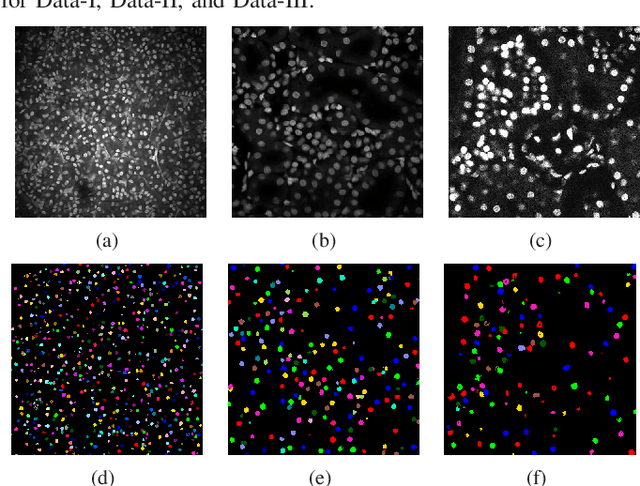
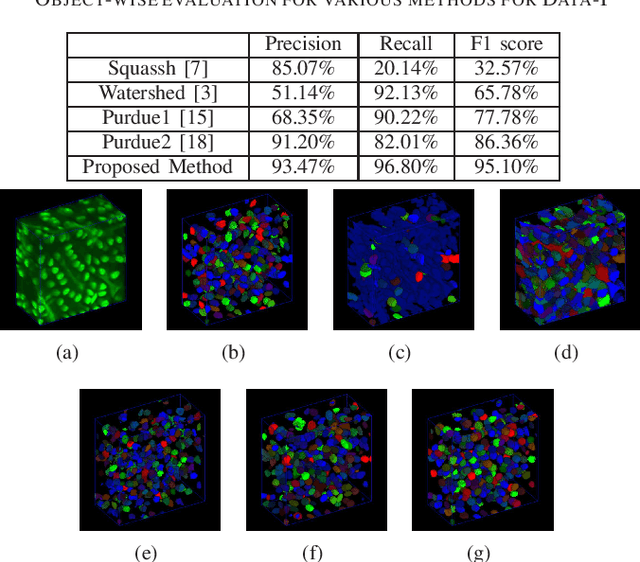
Abstract:Fluorescence microscopy is an essential tool for the analysis of 3D subcellular structures in tissue. An important step in the characterization of tissue involves nuclei segmentation. In this paper, a two-stage method for segmentation of nuclei using convolutional neural networks (CNNs) is described. In particular, since creating labeled volumes manually for training purposes is not practical due to the size and complexity of the 3D data sets, the paper describes a method for generating synthetic microscopy volumes based on a spatially constrained cycle-consistent adversarial network. The proposed method is tested on multiple real microscopy data sets and outperforms other commonly used segmentation techniques.
Single-View Food Portion Estimation: Learning Image-to-Energy Mappings Using Generative Adversarial Networks
May 23, 2018
Abstract:Due to the growing concern of chronic diseases and other health problems related to diet, there is a need to develop accurate methods to estimate an individual's food and energy intake. Measuring accurate dietary intake is an open research problem. In particular, accurate food portion estimation is challenging since the process of food preparation and consumption impose large variations on food shapes and appearances. In this paper, we present a food portion estimation method to estimate food energy (kilocalories) from food images using Generative Adversarial Networks (GAN). We introduce the concept of an "energy distribution" for each food image. To train the GAN, we design a food image dataset based on ground truth food labels and segmentation masks for each food image as well as energy information associated with the food image. Our goal is to learn the mapping of the food image to the food energy. We can then estimate food energy based on the energy distribution. We show that an average energy estimation error rate of 10.89% can be obtained by learning the image-to-energy mapping.
Three Dimensional Fluorescence Microscopy Image Synthesis and Segmentation
Apr 21, 2018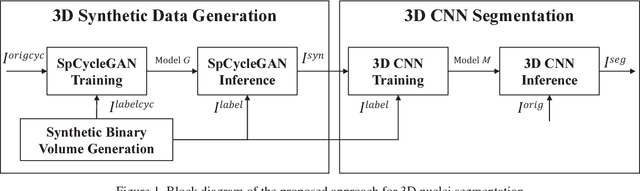
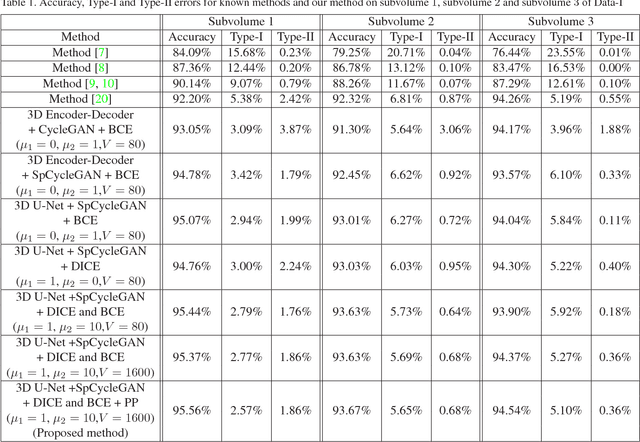
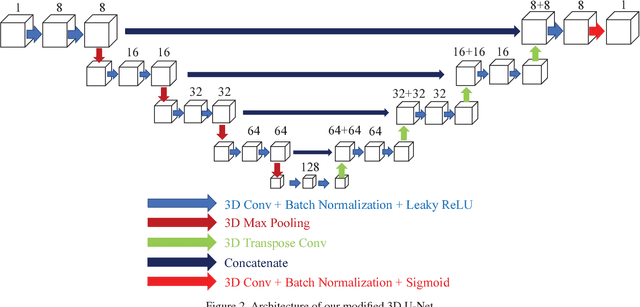
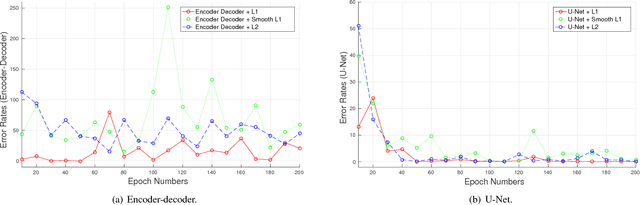
Abstract:Advances in fluorescence microscopy enable acquisition of 3D image volumes with better image quality and deeper penetration into tissue. Segmentation is a required step to characterize and analyze biological structures in the images and recent 3D segmentation using deep learning has achieved promising results. One issue is that deep learning techniques require a large set of groundtruth data which is impractical to annotate manually for large 3D microscopy volumes. This paper describes a 3D deep learning nuclei segmentation method using synthetic 3D volumes for training. A set of synthetic volumes and the corresponding groundtruth are generated using spatially constrained cycle-consistent adversarial networks. Segmentation results demonstrate that our proposed method is capable of segmenting nuclei successfully for various data sets.
Tubule segmentation of fluorescence microscopy images based on convolutional neural networks with inhomogeneity correction
Feb 10, 2018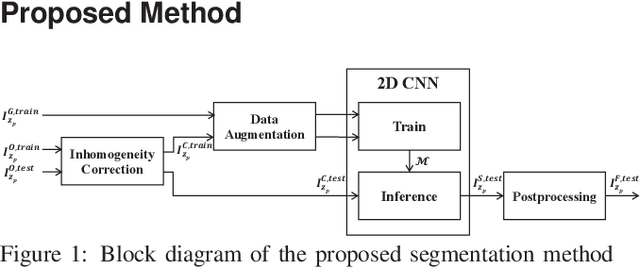
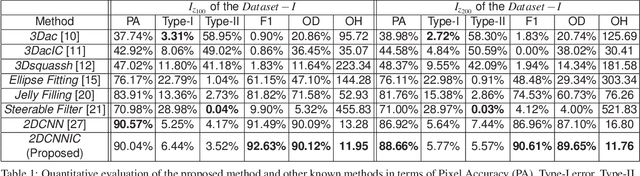
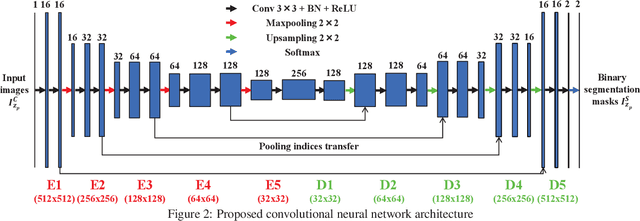

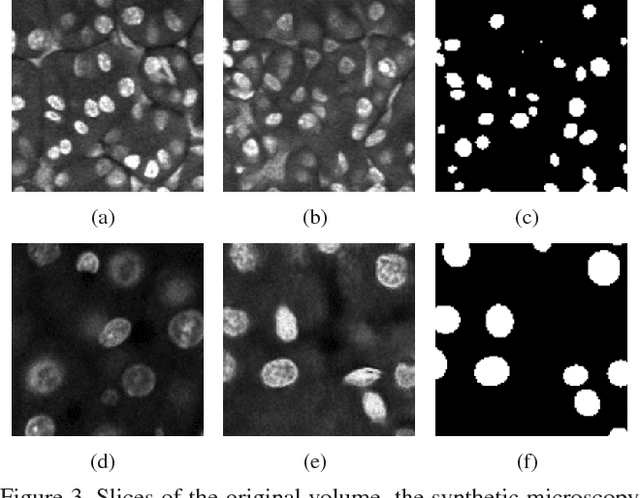
Abstract:Fluorescence microscopy has become a widely used tool for studying various biological structures of in vivo tissue or cells. However, quantitative analysis of these biological structures remains a challenge due to their complexity which is exacerbated by distortions caused by lens aberrations and light scattering. Moreover, manual quantification of such image volumes is an intractable and error-prone process, making the need for automated image analysis methods crucial. This paper describes a segmentation method for tubular structures in fluorescence microscopy images using convolutional neural networks with data augmentation and inhomogeneity correction. The segmentation results of the proposed method are visually and numerically compared with other microscopy segmentation methods. Experimental results indicate that the proposed method has better performance with correctly segmenting and identifying multiple tubular structures compared to other methods.
Texture Segmentation Based Video Compression Using Convolutional Neural Networks
Feb 08, 2018



Abstract:There has been a growing interest in using different approaches to improve the coding efficiency of modern video codec in recent years as demand for web-based video consumption increases. In this paper, we propose a model-based approach that uses texture analysis/synthesis to reconstruct blocks in texture regions of a video to achieve potential coding gains using the AV1 codec developed by the Alliance for Open Media (AOM). The proposed method uses convolutional neural networks to extract texture regions in a frame, which are then reconstructed using a global motion model. Our preliminary results show an increase in coding efficiency while maintaining satisfactory visual quality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge