David Joon Ho
Stain-invariant self supervised learning for histopathology image analysis
Nov 14, 2022
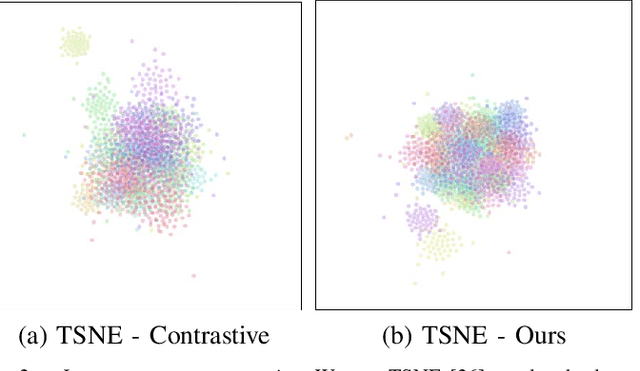

Abstract:We present a self-supervised algorithm for several classification tasks within hematoxylin and eosin (H&E) stained images of breast cancer. Our method is robust to stain variations inherent to the histology images acquisition process, which has limited the applicability of automated analysis tools. We address this problem by imposing constraints a learnt latent space which leverages stain normalization techniques during training. At every iteration, we select an image as a normalization target and generate a version of every image in the batch normalized to that target. We minimize the distance between the embeddings that correspond to the same image under different staining variations while maximizing the distance between other samples. We show that our method not only improves robustness to stain variations across multi-center data, but also classification performance through extensive experiments on various normalization targets and methods. Our method achieves the state-of-the-art performance on several publicly available breast cancer datasets ranging from tumor classification (CAMELYON17) and subtyping (BRACS) to HER2 status classification and treatment response prediction.
Deep Learning-Based Objective and Reproducible Osteosarcoma Chemotherapy Response Assessment and Outcome Prediction
Aug 09, 2022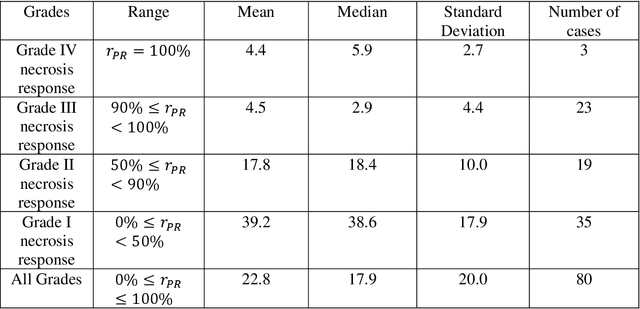
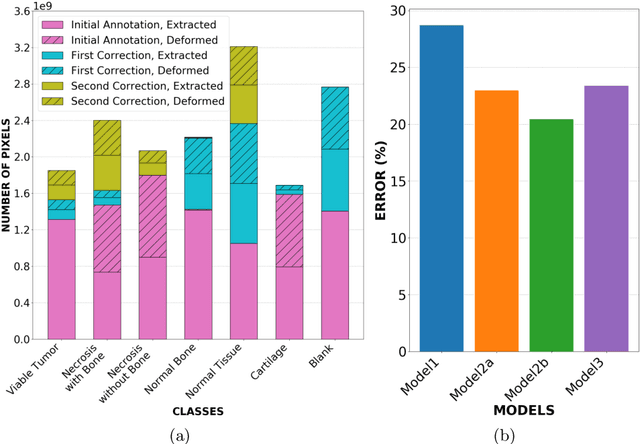
Abstract:Osteosarcoma is the most common primary bone cancer whose standard treatment includes pre-operative chemotherapy followed by resection. Chemotherapy response is used for predicting prognosis and further management of patients. Necrosis is routinely assessed post-chemotherapy from histology slides on resection specimens where necrosis ratio is defined as the ratio of necrotic tumor to overall tumor. Patients with necrosis ratio >=90% are known to have better outcome. Manual microscopic review of necrosis ratio from multiple glass slides is semi-quantitative and can have intra- and inter-observer variability. We propose an objective and reproducible deep learning-based approach to estimate necrosis ratio with outcome prediction from scanned hematoxylin and eosin whole slide images. We collected 103 osteosarcoma cases with 3134 WSIs to train our deep learning model, to validate necrosis ratio assessment, and to evaluate outcome prediction. We trained Deep Multi-Magnification Network to segment multiple tissue subtypes including viable tumor and necrotic tumor in pixel-level and to calculate case-level necrosis ratio from multiple WSIs. We showed necrosis ratio estimated by our segmentation model highly correlates with necrosis ratio from pathology reports manually assessed by experts where mean absolute differences for Grades IV (100%), III (>=90%), and II (>=50% and <90%) necrosis response are 4.4%, 4.5%, and 17.8%, respectively. We successfully stratified patients to predict overall survival with p=10^-6 and progression-free survival with p=0.012. Our reproducible approach without variability enabled us to tune cutoff thresholds, specifically for our model and our data set, to 80% for OS and 60% for PFS. Our study indicates deep learning can support pathologists as an objective tool to analyze osteosarcoma from histology for assessing treatment response and predicting patient outcome.
Deep Interactive Learning-based ovarian cancer segmentation of H&E-stained whole slide images to study morphological patterns of BRCA mutation
Mar 28, 2022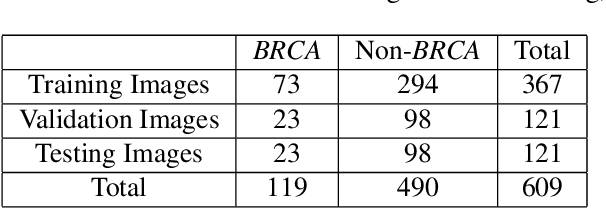
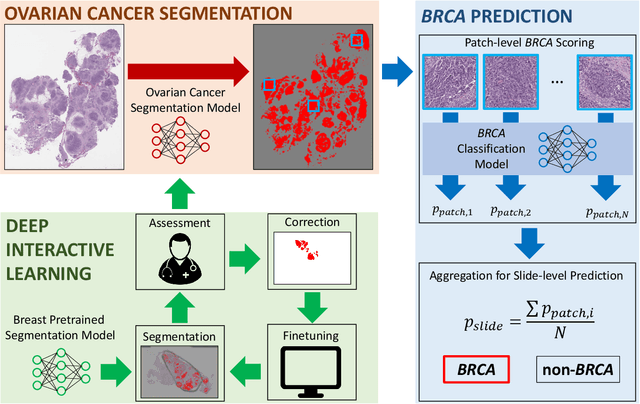
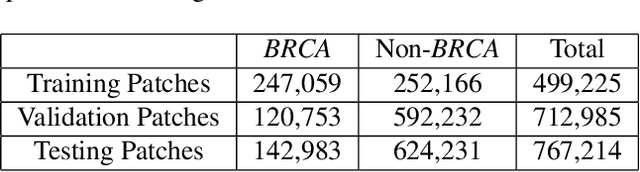
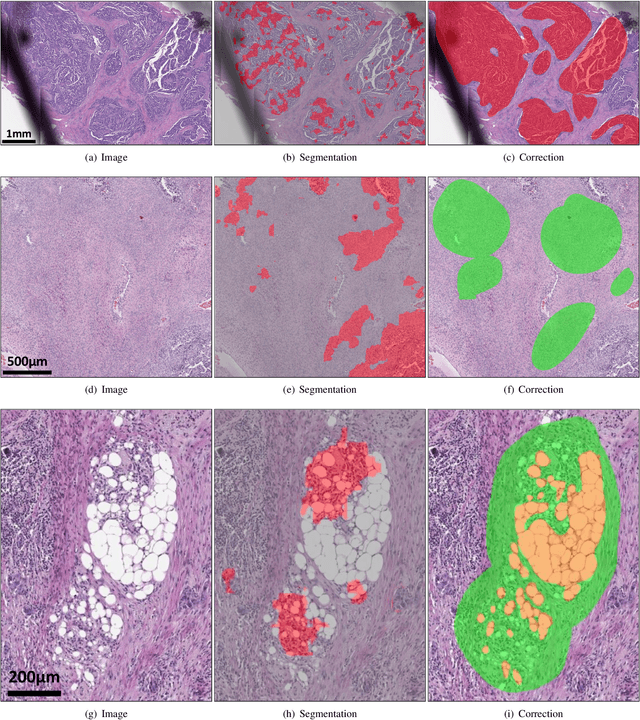
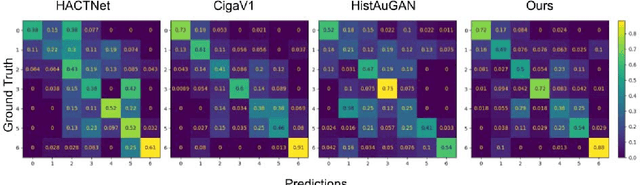
Abstract:Deep learning has been widely used to analyze digitized hematoxylin and eosin (H&E)-stained histopathology whole slide images. Automated cancer segmentation using deep learning can be used to diagnose malignancy and to find novel morphological patterns to predict molecular subtypes. To train pixel-wise cancer segmentation models, manual annotation from pathologists is generally a bottleneck due to its time-consuming nature. In this paper, we propose Deep Interactive Learning with a pretrained segmentation model from a different cancer type to reduce manual annotation time. Instead of annotating all pixels from cancer and non-cancer regions on giga-pixel whole slide images, an iterative process of annotating mislabeled regions from a segmentation model and training/finetuning the model with the additional annotation can reduce the time. Especially, employing a pretrained segmentation model can further reduce the time than starting annotation from scratch. We trained an accurate ovarian cancer segmentation model with a pretrained breast segmentation model by 3.5 hours of manual annotation which achieved intersection-over-union of 0.74, recall of 0.86, and precision of 0.84. With automatically extracted high-grade serous ovarian cancer patches, we attempted to train another deep learning model to predict BRCA mutation. The segmentation model and code have been released at https://github.com/MSKCC-Computational-Pathology/DMMN-ovary.
Deep Interactive Learning: An Efficient Labeling Approach for Deep Learning-Based Osteosarcoma Treatment Response Assessment
Jul 02, 2020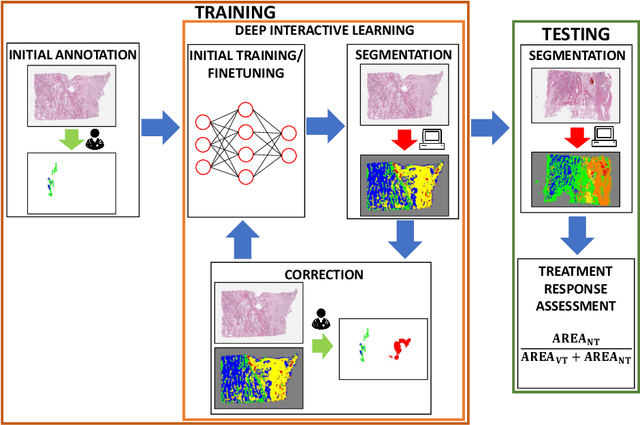
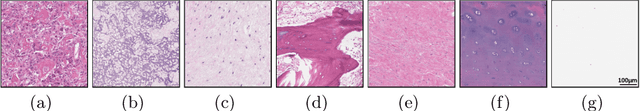
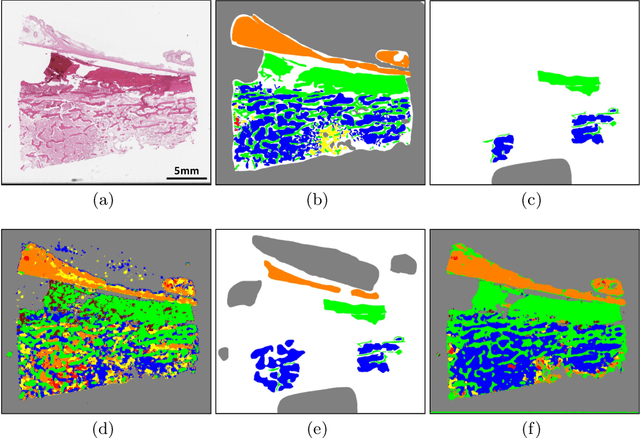
Abstract:Osteosarcoma is the most common malignant primary bone tumor. Standard treatment includes pre-operative chemotherapy followed by surgical resection. The response to treatment as measured by ratio of necrotic tumor area to overall tumor area is a known prognostic factor for overall survival. This assessment is currently done manually by pathologists by looking at glass slides under the microscope which may not be reproducible due to its subjective nature. Convolutional neural networks (CNNs) can be used for automated segmentation of viable and necrotic tumor on osteosarcoma whole slide images. One bottleneck for supervised learning is that large amounts of accurate annotations are required for training which is a time-consuming and expensive process. In this paper, we describe Deep Interactive Learning (DIaL) as an efficient labeling approach for training CNNs. After an initial labeling step is done, annotators only need to correct mislabeled regions from previous segmentation predictions to improve the CNN model until the satisfactory predictions are achieved. Our experiments show that our CNN model trained by only 7 hours of annotation using DIaL can successfully estimate ratios of necrosis within expected inter-observer variation rate for non-standardized manual surgical pathology task.
Deep Multi-Magnification Networks for Multi-Class Breast Cancer Image Segmentation
Oct 29, 2019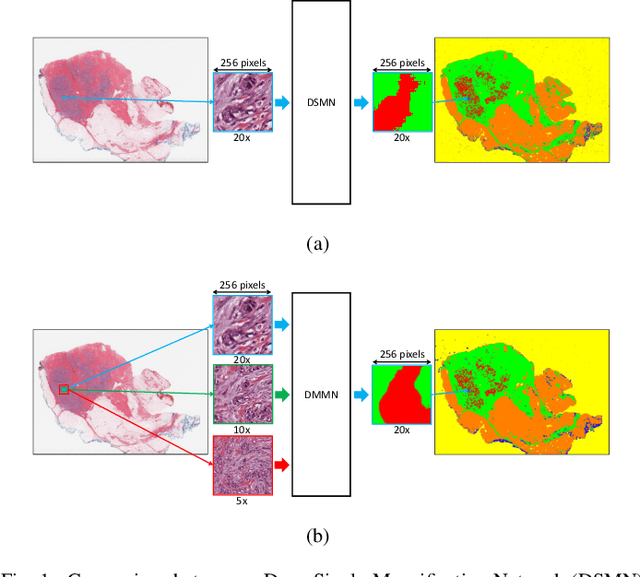
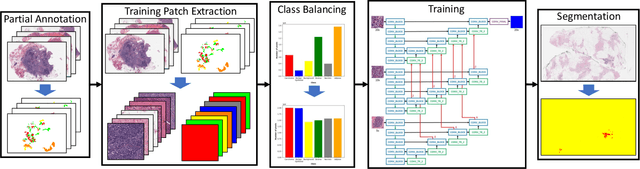
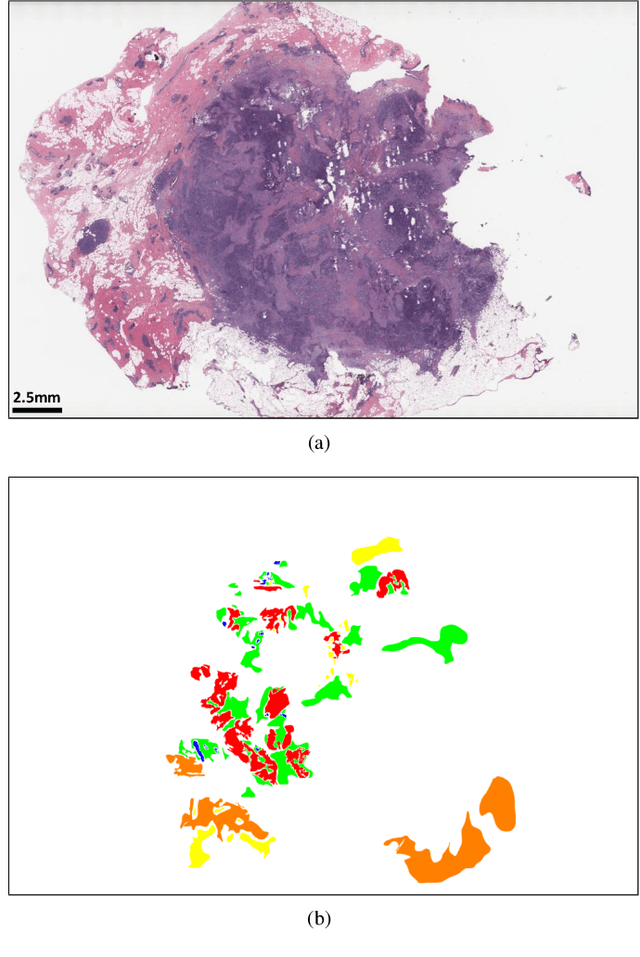
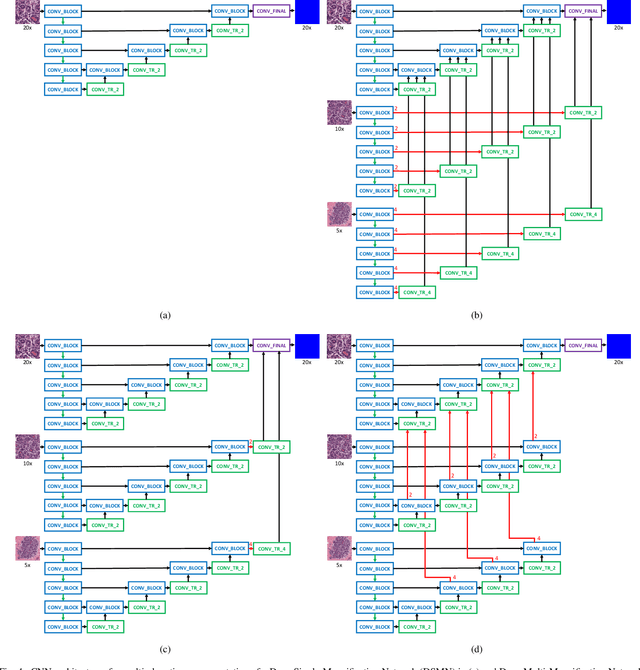
Abstract:Breast carcinoma is one of the most common cancers for women in the United States. Pathologic analysis of surgical excision specimens for breast carcinoma is important to evaluate the completeness of surgical excision and has implications for future treatment. This analysis is performed manually by pathologists reviewing histologic slides prepared from formalin-fixed tissue. Digital pathology has provided means to digitize the glass slides and generate whole slide images. Computational pathology enables whole slide images to be automatically analyzed to assist pathologists, especially with the advancement of deep learning. The whole slide images generally contain giga-pixels of data, so it is impractical to process the images at the whole-slide-level. Most of the current deep learning techniques process the images at the patch-level, but they may produce poor results by looking at individual patches with a narrow field-of-view at a single magnification. In this paper, we present Deep Multi-Magnification Networks (DMMNs) to resemble how pathologists analyze histologic slides using microscopes. Our multi-class tissue segmentation architecture processes a set of patches from multiple magnifications to make more accurate predictions. For our supervised training, we use partial annotations to reduce the burden of annotators. Our segmentation architecture with multi-encoder, multi-decoder, and multi-concatenation outperforms other segmentation architectures on breast datasets and can be used to facilitate pathologists' assessments of breast cancer.
Center-Extraction-Based Three Dimensional Nuclei Instance Segmentation of Fluorescence Microscopy Images
Sep 13, 2019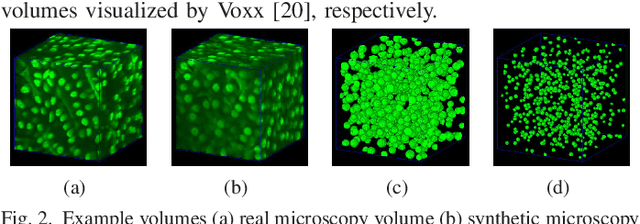
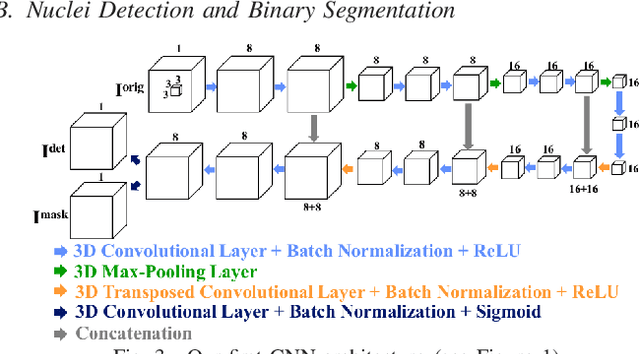
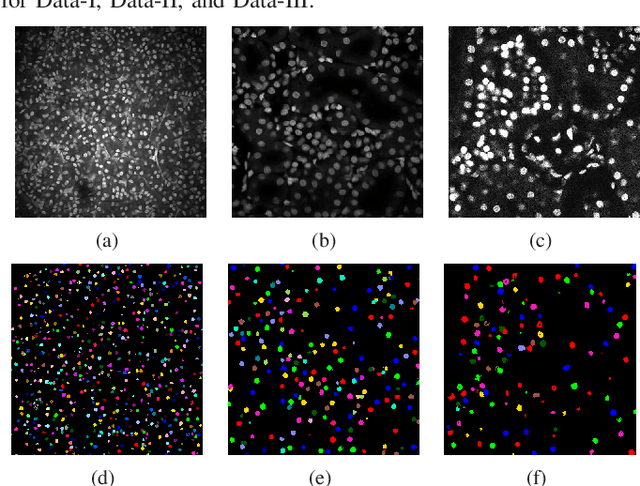
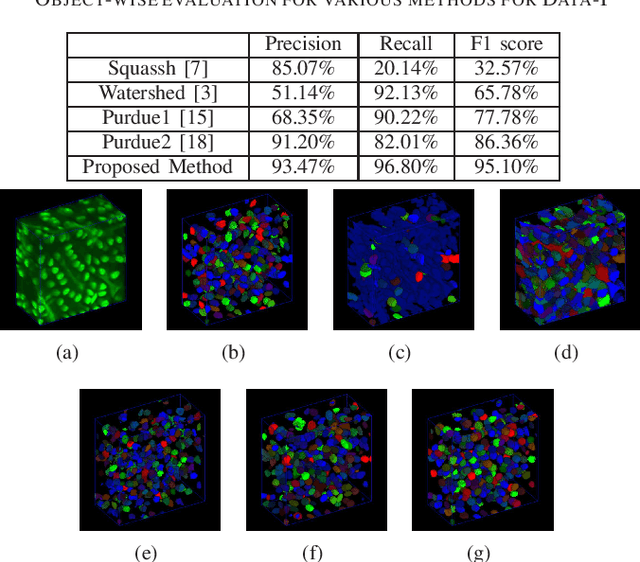
Abstract:Fluorescence microscopy is an essential tool for the analysis of 3D subcellular structures in tissue. An important step in the characterization of tissue involves nuclei segmentation. In this paper, a two-stage method for segmentation of nuclei using convolutional neural networks (CNNs) is described. In particular, since creating labeled volumes manually for training purposes is not practical due to the size and complexity of the 3D data sets, the paper describes a method for generating synthetic microscopy volumes based on a spatially constrained cycle-consistent adversarial network. The proposed method is tested on multiple real microscopy data sets and outperforms other commonly used segmentation techniques.
Three Dimensional Fluorescence Microscopy Image Synthesis and Segmentation
Apr 21, 2018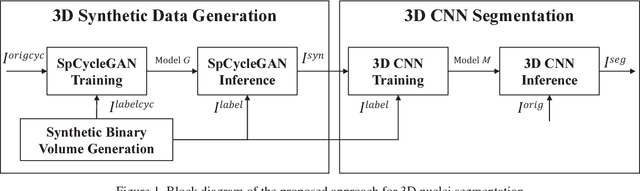
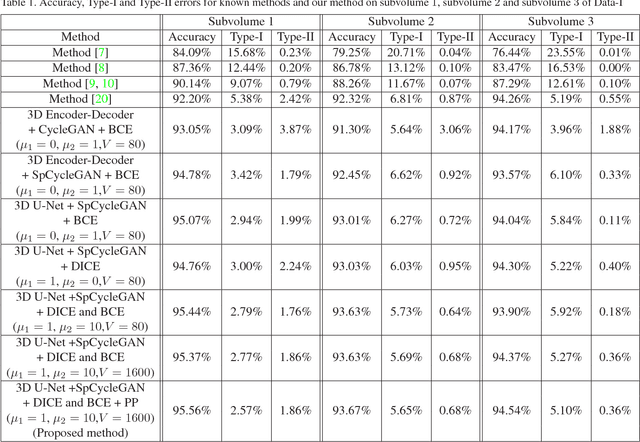
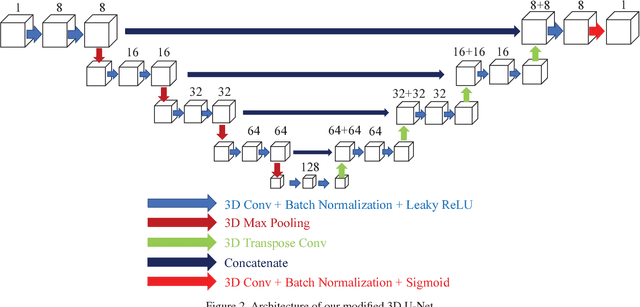
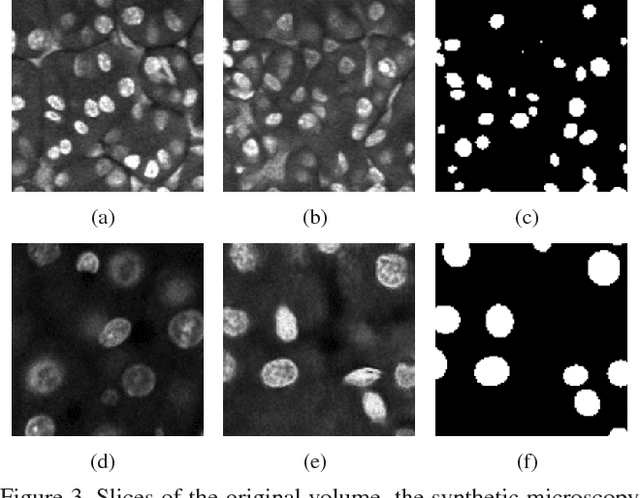
Abstract:Advances in fluorescence microscopy enable acquisition of 3D image volumes with better image quality and deeper penetration into tissue. Segmentation is a required step to characterize and analyze biological structures in the images and recent 3D segmentation using deep learning has achieved promising results. One issue is that deep learning techniques require a large set of groundtruth data which is impractical to annotate manually for large 3D microscopy volumes. This paper describes a 3D deep learning nuclei segmentation method using synthetic 3D volumes for training. A set of synthetic volumes and the corresponding groundtruth are generated using spatially constrained cycle-consistent adversarial networks. Segmentation results demonstrate that our proposed method is capable of segmenting nuclei successfully for various data sets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge