Saad Nadeem
Rethinking Histology Slide Digitization Workflows for Low-Resource Settings
May 13, 2024



Abstract:Histology slide digitization is becoming essential for telepathology (remote consultation), knowledge sharing (education), and using the state-of-the-art artificial intelligence algorithms (augmented/automated end-to-end clinical workflows). However, the cumulative costs of digital multi-slide high-speed brightfield scanners, cloud/on-premises storage, and personnel (IT and technicians) make the current slide digitization workflows out-of-reach for limited-resource settings, further widening the health equity gap; even single-slide manual scanning commercial solutions are costly due to hardware requirements (high-resolution cameras, high-spec PC/workstation, and support for only high-end microscopes). In this work, we present a new cloud slide digitization workflow for creating scanner-quality whole-slide images (WSIs) from uploaded low-quality videos, acquired from cheap and inexpensive microscopes with built-in cameras. Specifically, we present a pipeline to create stitched WSIs while automatically deblurring out-of-focus regions, upsampling input 10X images to 40X resolution, and reducing brightness/contrast and light-source illumination variations. We demonstrate the WSI creation efficacy from our workflow on World Health Organization-declared neglected tropical disease, Cutaneous Leishmaniasis (prevalent only in the poorest regions of the world and only diagnosed by sub-specialist dermatopathologists, rare in poor countries), as well as other common pathologies on core biopsies of breast, liver, duodenum, stomach and lymph node. The code and pretrained models will be accessible via our GitHub (https://github.com/nadeemlab/DeepLIIF), and the cloud platform will be available at https://deepliif.org for uploading microscope videos and downloading/viewing WSIs with shareable links (no sign-in required) for telepathology and knowledge sharing.
RT-GAN: Recurrent Temporal GAN for Adding Lightweight Temporal Consistency to Frame-Based Domain Translation Approaches
Oct 02, 2023Abstract:While developing new unsupervised domain translation methods for endoscopy videos, it is typical to start with approaches that initially work for individual frames without temporal consistency. Once an individual-frame model has been finalized, additional contiguous frames are added with a modified deep learning architecture to train a new model for temporal consistency. This transition to temporally-consistent deep learning models, however, requires significantly more computational and memory resources for training. In this paper, we present a lightweight solution with a tunable temporal parameter, RT-GAN (Recurrent Temporal GAN), for adding temporal consistency to individual frame-based approaches that reduces training requirements by a factor of 5. We demonstrate the effectiveness of our approach on two challenging use cases in colonoscopy: haustral fold segmentation (indicative of missed surface) and realistic colonoscopy simulator video generation. The datasets, accompanying code, and pretrained models will be made available at \url{https://github.com/nadeemlab/CEP}.
An AI-Ready Multiplex Staining Dataset for Reproducible and Accurate Characterization of Tumor Immune Microenvironment
May 25, 2023



Abstract:We introduce a new AI-ready computational pathology dataset containing restained and co-registered digitized images from eight head-and-neck squamous cell carcinoma patients. Specifically, the same tumor sections were stained with the expensive multiplex immunofluorescence (mIF) assay first and then restained with cheaper multiplex immunohistochemistry (mIHC). This is a first public dataset that demonstrates the equivalence of these two staining methods which in turn allows several use cases; due to the equivalence, our cheaper mIHC staining protocol can offset the need for expensive mIF staining/scanning which requires highly-skilled lab technicians. As opposed to subjective and error-prone immune cell annotations from individual pathologists (disagreement > 50%) to drive SOTA deep learning approaches, this dataset provides objective immune and tumor cell annotations via mIF/mIHC restaining for more reproducible and accurate characterization of tumor immune microenvironment (e.g. for immunotherapy). We demonstrate the effectiveness of this dataset in three use cases: (1) IHC quantification of CD3/CD8 tumor-infiltrating lymphocytes via style transfer, (2) virtual translation of cheap mIHC stains to more expensive mIF stains, and (3) virtual tumor/immune cellular phenotyping on standard hematoxylin images. The dataset is available at \url{https://github.com/nadeemlab/DeepLIIF}.
RMSim: Controlled Respiratory Motion Simulation on Static Patient Scans
Jan 26, 2023Abstract:This work aims to generate realistic anatomical deformations from static patient scans. Specifically, we present a method to generate these deformations/augmentations via deep learning driven respiratory motion simulation that provides the ground truth for validating deformable image registration (DIR) algorithms and driving more accurate deep learning based DIR. We present a novel 3D Seq2Seq deep learning respiratory motion simulator (RMSim) that learns from 4D-CT images and predicts future breathing phases given a static CT image. The predicted respiratory patterns, represented by time-varying displacement vector fields (DVFs) at different breathing phases, are modulated through auxiliary inputs of 1D breathing traces so that a larger amplitude in the trace results in more significant predicted deformation. Stacked 3D-ConvLSTMs are used to capture the spatial-temporal respiration patterns. Training loss includes a smoothness loss in the DVF and mean-squared error between the predicted and ground truth phase images. A spatial transformer deforms the static CT with the predicted DVF to generate the predicted phase image. 10-phase 4D-CTs of 140 internal patients were used to train and test RMSim. The trained RMSim was then used to augment a public DIR challenge dataset for training VoxelMorph to show the effectiveness of RMSim-generated deformation augmentation. We validated our RMSim output with both private and public benchmark datasets (healthy and cancer patients). The proposed approach can be used for validating DIR algorithms as well as for patient-specific augmentations to improve deep learning DIR algorithms. The code, pretrained models, and augmented DIR validation datasets will be released at https://github.com/nadeemlab/SeqX2Y.
Stain-invariant self supervised learning for histopathology image analysis
Nov 14, 2022
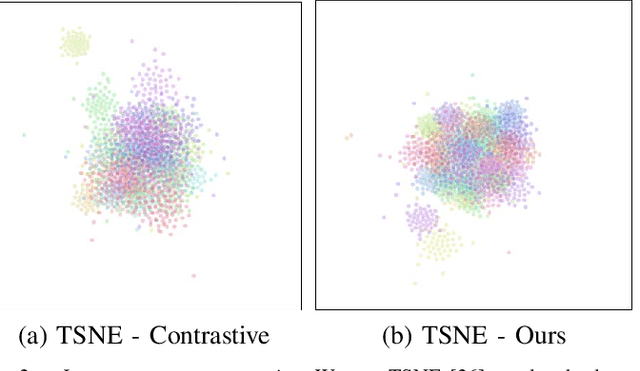

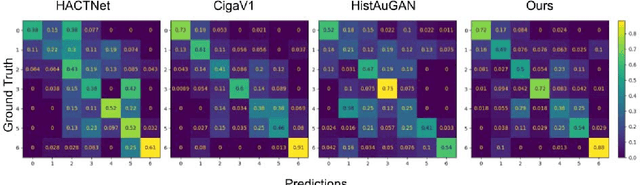
Abstract:We present a self-supervised algorithm for several classification tasks within hematoxylin and eosin (H&E) stained images of breast cancer. Our method is robust to stain variations inherent to the histology images acquisition process, which has limited the applicability of automated analysis tools. We address this problem by imposing constraints a learnt latent space which leverages stain normalization techniques during training. At every iteration, we select an image as a normalization target and generate a version of every image in the batch normalized to that target. We minimize the distance between the embeddings that correspond to the same image under different staining variations while maximizing the distance between other samples. We show that our method not only improves robustness to stain variations across multi-center data, but also classification performance through extensive experiments on various normalization targets and methods. Our method achieves the state-of-the-art performance on several publicly available breast cancer datasets ranging from tumor classification (CAMELYON17) and subtyping (BRACS) to HER2 status classification and treatment response prediction.
Domain Knowledge Driven 3D Dose Prediction Using Moment-Based Loss Function
Jul 07, 2022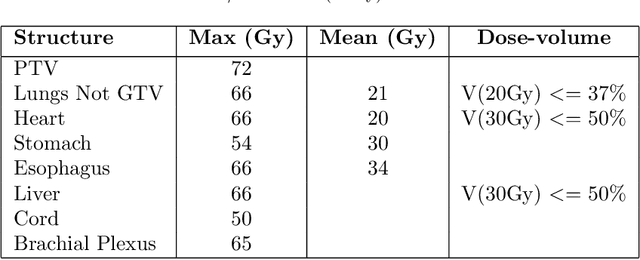
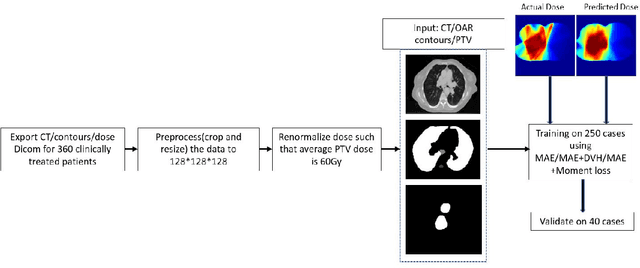
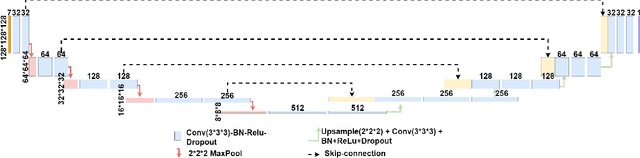
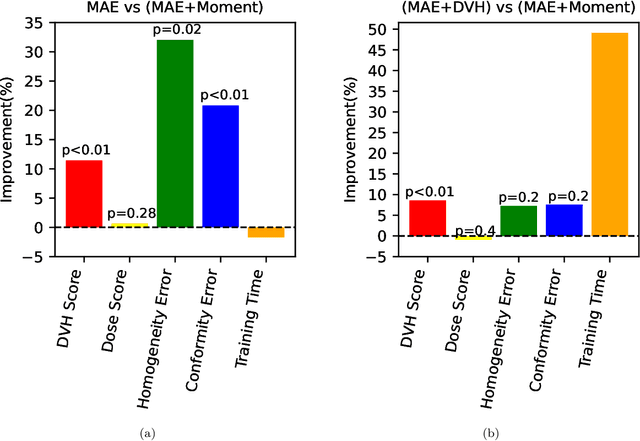
Abstract:Dose volume histogram (DVH) metrics are widely accepted evaluation criteria in the clinic. However, incorporating these metrics into deep learning dose prediction models is challenging due to their non-convexity and non-differentiability. We propose a novel moment-based loss function for predicting 3D dose distribution for the challenging conventional lung intensity modulated radiation therapy (IMRT) plans. The moment-based loss function is convex and differentiable and can easily incorporate DVH metrics in any deep learning framework without computational overhead. The moments can also be customized to reflect the clinical priorities in 3D dose prediction. For instance, using high-order moments allows better prediction in high-dose areas for serial structures. We used a large dataset of 360 (240 for training, 50 for validation and 70 for testing) conventional lung patients with 2Gy $\times$ 30 fractions to train the deep learning (DL) model using clinically treated plans at our institution. We trained a UNet like CNN architecture using computed tomography (CT), planning target volume (PTV) and organ-at-risk contours (OAR) as input to infer corresponding voxel-wise 3D dose distribution. We evaluated three different loss functions: (1) The popular Mean Absolute Error (MAE) Loss, (2) the recently developed MAE + DVH Loss, and (3) the proposed MAE + Moments Loss. The quality of the predictions was compared using different DVH metrics as well as dose-score and DVH-score, recently introduced by the AAPM knowledge-based planning grand challenge. Model with (MAE + Moment) loss function outperformed the model with MAE loss by significantly improving the DVH-score (11%, p$<$0.01) while having similar computational cost. It also outperformed the model trained with (MAE+DVH) by significantly improving the computational cost (48%) and the DVH-score (8%, p$<$0.01).
CLTS-GAN: Color-Lighting-Texture-Specular Reflection Augmentation for Colonoscopy
Jun 29, 2022



Abstract:Automated analysis of optical colonoscopy (OC) video frames (to assist endoscopists during OC) is challenging due to variations in color, lighting, texture, and specular reflections. Previous methods either remove some of these variations via preprocessing (making pipelines cumbersome) or add diverse training data with annotations (but expensive and time-consuming). We present CLTS-GAN, a new deep learning model that gives fine control over color, lighting, texture, and specular reflection synthesis for OC video frames. We show that adding these colonoscopy-specific augmentations to the training data can improve state-of-the-art polyp detection/segmentation methods as well as drive next generation of OC simulators for training medical students. The code and pre-trained models for CLTS-GAN are available on Computational Endoscopy Platform GitHub (https://github.com/nadeemlab/CEP).
CIRDataset: A large-scale Dataset for Clinically-Interpretable lung nodule Radiomics and malignancy prediction
Jun 29, 2022



Abstract:Spiculations/lobulations, sharp/curved spikes on the surface of lung nodules, are good predictors of lung cancer malignancy and hence, are routinely assessed and reported by radiologists as part of the standardized Lung-RADS clinical scoring criteria. Given the 3D geometry of the nodule and 2D slice-by-slice assessment by radiologists, manual spiculation/lobulation annotation is a tedious task and thus no public datasets exist to date for probing the importance of these clinically-reported features in the SOTA malignancy prediction algorithms. As part of this paper, we release a large-scale Clinically-Interpretable Radiomics Dataset, CIRDataset, containing 956 radiologist QA/QC'ed spiculation/lobulation annotations on segmented lung nodules from two public datasets, LIDC-IDRI (N=883) and LUNGx (N=73). We also present an end-to-end deep learning model based on multi-class Voxel2Mesh extension to segment nodules (while preserving spikes), classify spikes (sharp/spiculation and curved/lobulation), and perform malignancy prediction. Previous methods have performed malignancy prediction for LIDC and LUNGx datasets but without robust attribution to any clinically reported/actionable features (due to known hyperparameter sensitivity issues with general attribution schemes). With the release of this comprehensively-annotated CIRDataset and end-to-end deep learning baseline, we hope that malignancy prediction methods can validate their explanations, benchmark against our baseline, and provide clinically-actionable insights. Dataset, code, pretrained models, and docker containers are available at https://github.com/nadeemlab/CIR.
DeepLIIF: An Online Platform for Quantification of Clinical Pathology Slides
Apr 09, 2022



Abstract:In the clinic, resected tissue samples are stained with Hematoxylin-and-Eosin (H&E) and/or Immunhistochemistry (IHC) stains and presented to the pathologists on glass slides or as digital scans for diagnosis and assessment of disease progression. Cell-level quantification, e.g. in IHC protein expression scoring, can be extremely inefficient and subjective. We present DeepLIIF (https://deepliif.org), a first free online platform for efficient and reproducible IHC scoring. DeepLIIF outperforms current state-of-the-art approaches (relying on manual error-prone annotations) by virtually restaining clinical IHC slides with more informative multiplex immunofluorescence staining. Our DeepLIIF cloud-native platform supports (1) more than 150 proprietary/non-proprietary input formats via the Bio-Formats standard, (2) interactive adjustment, visualization, and downloading of the IHC quantification results and the accompanying restained images, (3) consumption of an exposed workflow API programmatically or through interactive plugins for open source whole slide image viewers such as QuPath/ImageJ, and (4) auto scaling to efficiently scale GPU resources based on user demand.
FoldIt: Haustral Folds Detection and Segmentation in Colonoscopy Videos
Jun 23, 2021



Abstract:Haustral folds are colon wall protrusions implicated for high polyp miss rate during optical colonoscopy procedures. If segmented accurately, haustral folds can allow for better estimation of missed surface and can also serve as valuable landmarks for registering pre-treatment virtual (CT) and optical colonoscopies, to guide navigation towards the anomalies found in pre-treatment scans. We present a novel generative adversarial network, FoldIt, for feature-consistent image translation of optical colonoscopy videos to virtual colonoscopy renderings with haustral fold overlays. A new transitive loss is introduced in order to leverage ground truth information between haustral fold annotations and virtual colonoscopy renderings. We demonstrate the effectiveness of our model on real challenging optical colonoscopy videos as well as on textured virtual colonoscopy videos with clinician-verified haustral fold annotations. All code and scripts to reproduce the experiments of this paper will be made available via our Computational Endoscopy Platform at https://github.com/nadeemlab/CEP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge