Shawn Mathew
RT-GAN: Recurrent Temporal GAN for Adding Lightweight Temporal Consistency to Frame-Based Domain Translation Approaches
Oct 02, 2023Abstract:While developing new unsupervised domain translation methods for endoscopy videos, it is typical to start with approaches that initially work for individual frames without temporal consistency. Once an individual-frame model has been finalized, additional contiguous frames are added with a modified deep learning architecture to train a new model for temporal consistency. This transition to temporally-consistent deep learning models, however, requires significantly more computational and memory resources for training. In this paper, we present a lightweight solution with a tunable temporal parameter, RT-GAN (Recurrent Temporal GAN), for adding temporal consistency to individual frame-based approaches that reduces training requirements by a factor of 5. We demonstrate the effectiveness of our approach on two challenging use cases in colonoscopy: haustral fold segmentation (indicative of missed surface) and realistic colonoscopy simulator video generation. The datasets, accompanying code, and pretrained models will be made available at \url{https://github.com/nadeemlab/CEP}.
CLTS-GAN: Color-Lighting-Texture-Specular Reflection Augmentation for Colonoscopy
Jun 29, 2022



Abstract:Automated analysis of optical colonoscopy (OC) video frames (to assist endoscopists during OC) is challenging due to variations in color, lighting, texture, and specular reflections. Previous methods either remove some of these variations via preprocessing (making pipelines cumbersome) or add diverse training data with annotations (but expensive and time-consuming). We present CLTS-GAN, a new deep learning model that gives fine control over color, lighting, texture, and specular reflection synthesis for OC video frames. We show that adding these colonoscopy-specific augmentations to the training data can improve state-of-the-art polyp detection/segmentation methods as well as drive next generation of OC simulators for training medical students. The code and pre-trained models for CLTS-GAN are available on Computational Endoscopy Platform GitHub (https://github.com/nadeemlab/CEP).
NeuRegenerate: A Framework for Visualizing Neurodegeneration
Feb 02, 2022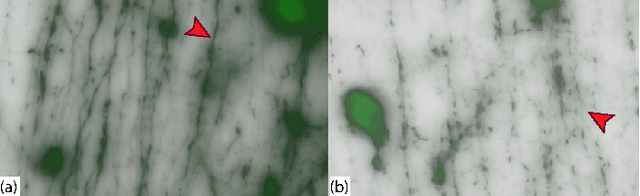
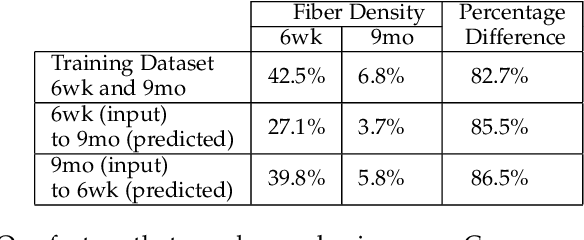
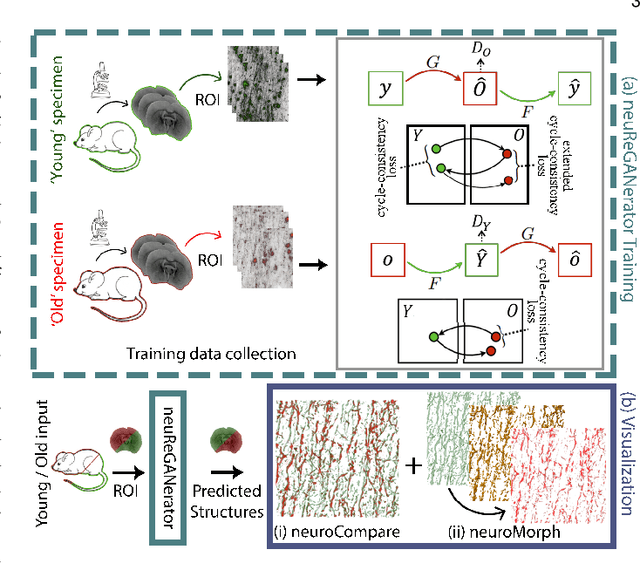
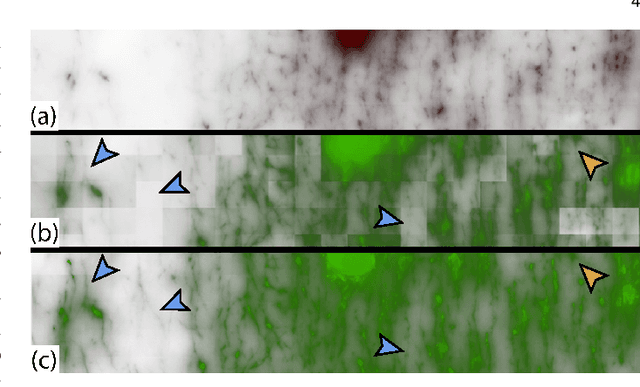
Abstract:Recent advances in high-resolution microscopy have allowed scientists to better understand the underlying brain connectivity. However, due to the limitation that biological specimens can only be imaged at a single timepoint, studying changes to neural projections is limited to general observations using population analysis. In this paper, we introduce NeuRegenerate, a novel end-to-end framework for the prediction and visualization of changes in neural fiber morphology within a subject, for specified age-timepoints.To predict projections, we present neuReGANerator, a deep-learning network based on cycle-consistent generative adversarial network (cycleGAN) that translates features of neuronal structures in a region, across age-timepoints, for large brain microscopy volumes. We improve the reconstruction quality of neuronal structures by implementing a density multiplier and a new loss function, called the hallucination loss.Moreover, to alleviate artifacts that occur due to tiling of large input volumes, we introduce a spatial-consistency module in the training pipeline of neuReGANerator. We show that neuReGANerator has a reconstruction accuracy of 94% in predicting neuronal structures. Finally, to visualize the predicted change in projections, NeuRegenerate offers two modes: (1) neuroCompare to simultaneously visualize the difference in the structures of the neuronal projections, across the age timepoints, and (2) neuroMorph, a vesselness-based morphing technique to interactively visualize the transformation of the structures from one age-timepoint to the other. Our framework is designed specifically for volumes acquired using wide-field microscopy. We demonstrate our framework by visualizing the structural changes in neuronal fibers within the cholinergic system of the mouse brain between a young and old specimen.
FoldIt: Haustral Folds Detection and Segmentation in Colonoscopy Videos
Jun 23, 2021



Abstract:Haustral folds are colon wall protrusions implicated for high polyp miss rate during optical colonoscopy procedures. If segmented accurately, haustral folds can allow for better estimation of missed surface and can also serve as valuable landmarks for registering pre-treatment virtual (CT) and optical colonoscopies, to guide navigation towards the anomalies found in pre-treatment scans. We present a novel generative adversarial network, FoldIt, for feature-consistent image translation of optical colonoscopy videos to virtual colonoscopy renderings with haustral fold overlays. A new transitive loss is introduced in order to leverage ground truth information between haustral fold annotations and virtual colonoscopy renderings. We demonstrate the effectiveness of our model on real challenging optical colonoscopy videos as well as on textured virtual colonoscopy videos with clinician-verified haustral fold annotations. All code and scripts to reproduce the experiments of this paper will be made available via our Computational Endoscopy Platform at https://github.com/nadeemlab/CEP.
Visualizing Missing Surfaces In Colonoscopy Videos using Shared Latent Space Representations
Jan 18, 2021



Abstract:Optical colonoscopy (OC), the most prevalent colon cancer screening tool, has a high miss rate due to a number of factors, including the geometry of the colon (haustral fold and sharp bends occlusions), endoscopist inexperience or fatigue, endoscope field of view, etc. We present a framework to visualize the missed regions per-frame during the colonoscopy, and provides a workable clinical solution. Specifically, we make use of 3D reconstructed virtual colonoscopy (VC) data and the insight that VC and OC share the same underlying geometry but differ in color, texture and specular reflections, embedded in the OC domain. A lossy unpaired image-to-image translation model is introduced with enforced shared latent space for OC and VC. This shared latent space captures the geometric information while deferring the color, texture, and specular information creation to additional Gaussian noise input. This additional noise input can be utilized to generate one-to-many mappings from VC to OC and OC to OC.
Augmenting Colonoscopy using Extended and Directional CycleGAN for Lossy Image Translation
Mar 27, 2020



Abstract:Colorectal cancer screening modalities, such as optical colonoscopy (OC) and virtual colonoscopy (VC), are critical for diagnosing and ultimately removing polyps (precursors of colon cancer). The non-invasive VC is normally used to inspect a 3D reconstructed colon (from CT scans) for polyps and if found, the OC procedure is performed to physically traverse the colon via endoscope and remove these polyps. In this paper, we present a deep learning framework, Extended and Directional CycleGAN, for lossy unpaired image-to-image translation between OC and VC to augment OC video sequences with scale-consistent depth information from VC, and augment VC with patient-specific textures, color and specular highlights from OC (e.g, for realistic polyp synthesis). Both OC and VC contain structural information, but it is obscured in OC by additional patient-specific texture and specular highlights, hence making the translation from OC to VC lossy. The existing CycleGAN approaches do not handle lossy transformations. To address this shortcoming, we introduce an extended cycle consistency loss, which compares the geometric structures from OC in the VC domain. This loss removes the need for the CycleGAN to embed OC information in the VC domain. To handle a stronger removal of the textures and lighting, a Directional Discriminator is introduced to differentiate the direction of translation (by creating paired information for the discriminator), as opposed to the standard CycleGAN which is direction-agnostic. Combining the extended cycle consistency loss and the Directional Discriminator, we show state-of-the-art results on scale-consistent depth inference for phantom, textured VC and for real polyp and normal colon video sequences. We also present results for realistic pendunculated and flat polyp synthesis from bumps introduced in 3D VC models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge