Alexandre Tiard
Stain-invariant self supervised learning for histopathology image analysis
Nov 14, 2022
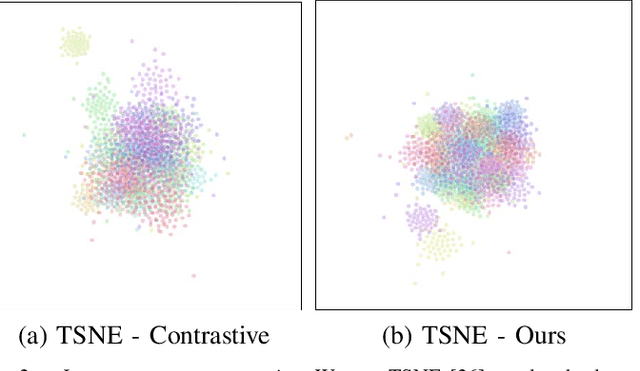

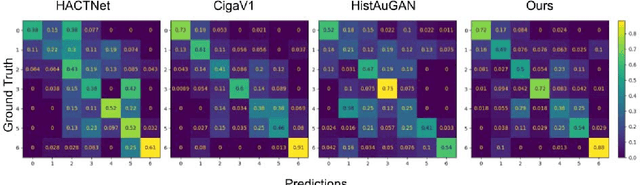
Abstract:We present a self-supervised algorithm for several classification tasks within hematoxylin and eosin (H&E) stained images of breast cancer. Our method is robust to stain variations inherent to the histology images acquisition process, which has limited the applicability of automated analysis tools. We address this problem by imposing constraints a learnt latent space which leverages stain normalization techniques during training. At every iteration, we select an image as a normalization target and generate a version of every image in the batch normalized to that target. We minimize the distance between the embeddings that correspond to the same image under different staining variations while maximizing the distance between other samples. We show that our method not only improves robustness to stain variations across multi-center data, but also classification performance through extensive experiments on various normalization targets and methods. Our method achieves the state-of-the-art performance on several publicly available breast cancer datasets ranging from tumor classification (CAMELYON17) and subtyping (BRACS) to HER2 status classification and treatment response prediction.
Small Lesion Segmentation in Brain MRIs with Subpixel Embedding
Sep 18, 2021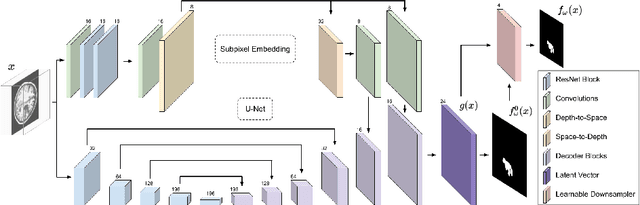

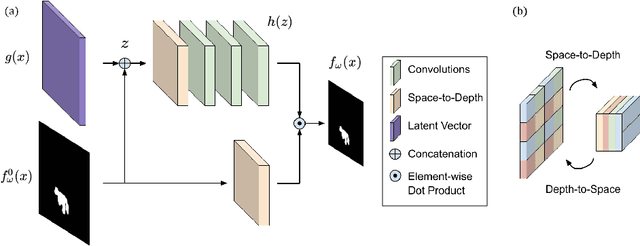
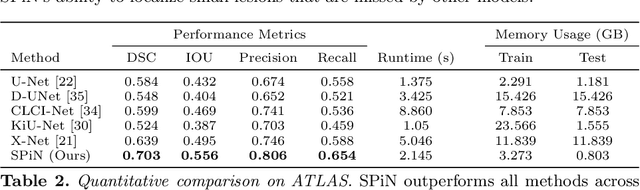
Abstract:We present a method to segment MRI scans of the human brain into ischemic stroke lesion and normal tissues. We propose a neural network architecture in the form of a standard encoder-decoder where predictions are guided by a spatial expansion embedding network. Our embedding network learns features that can resolve detailed structures in the brain without the need for high-resolution training images, which are often unavailable and expensive to acquire. Alternatively, the encoder-decoder learns global structures by means of striding and max pooling. Our embedding network complements the encoder-decoder architecture by guiding the decoder with fine-grained details lost to spatial downsampling during the encoder stage. Unlike previous works, our decoder outputs at 2 times the input resolution, where a single pixel in the input resolution is predicted by four neighboring subpixels in our output. To obtain the output at the original scale, we propose a learnable downsampler (as opposed to hand-crafted ones e.g. bilinear) that combines subpixel predictions. Our approach improves the baseline architecture by approximately 11.7% and achieves the state of the art on the ATLAS public benchmark dataset with a smaller memory footprint and faster runtime than the best competing method. Our source code has been made available at: https://github.com/alexklwong/subpixel-embedding-segmentation.
Unsupervised Classification in Hyperspectral Imagery with Nonlocal Total Variation and Primal-Dual Hybrid Gradient Algorithm
Feb 13, 2017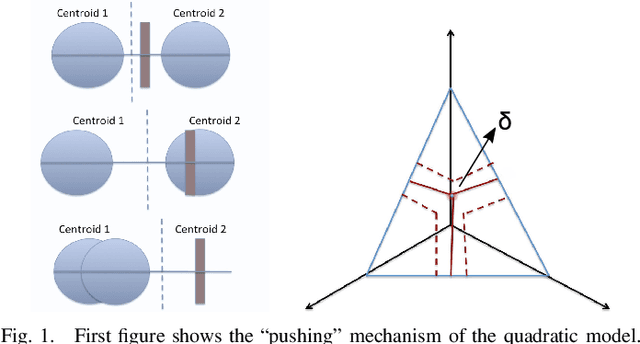
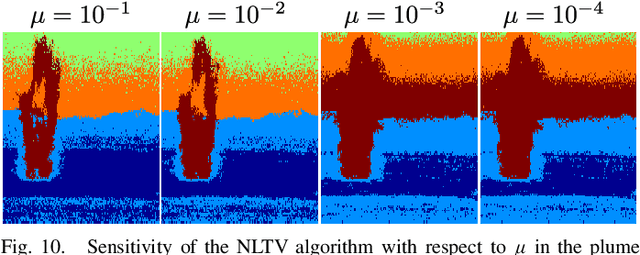

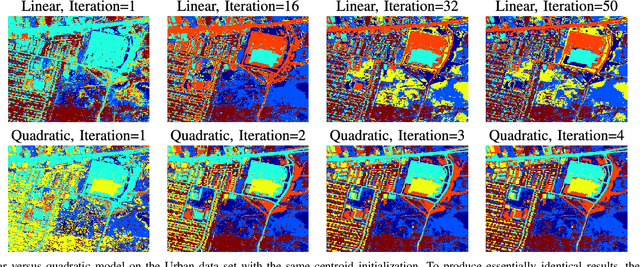
Abstract:In this paper, a graph-based nonlocal total variation method (NLTV) is proposed for unsupervised classification of hyperspectral images (HSI). The variational problem is solved by the primal-dual hybrid gradient (PDHG) algorithm. By squaring the labeling function and using a stable simplex clustering routine, an unsupervised clustering method with random initialization can be implemented. The effectiveness of this proposed algorithm is illustrated on both synthetic and real-world HSI, and numerical results show that the proposed algorithm outperforms other standard unsupervised clustering methods such as spherical K-means, nonnegative matrix factorization (NMF), and the graph-based Merriman-Bence-Osher (MBO) scheme.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge