Navdeep Dahiya
Domain Knowledge Driven 3D Dose Prediction Using Moment-Based Loss Function
Jul 07, 2022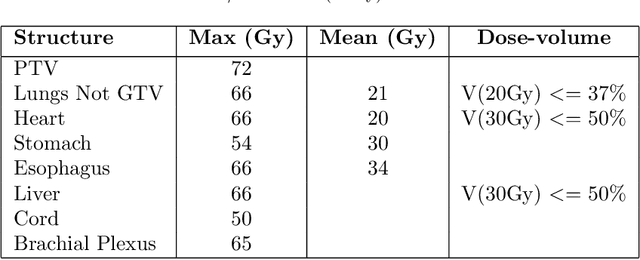
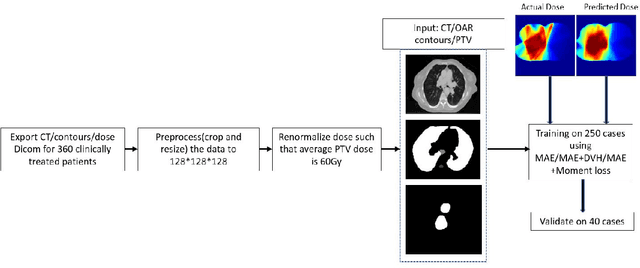
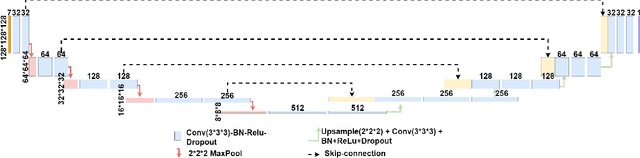
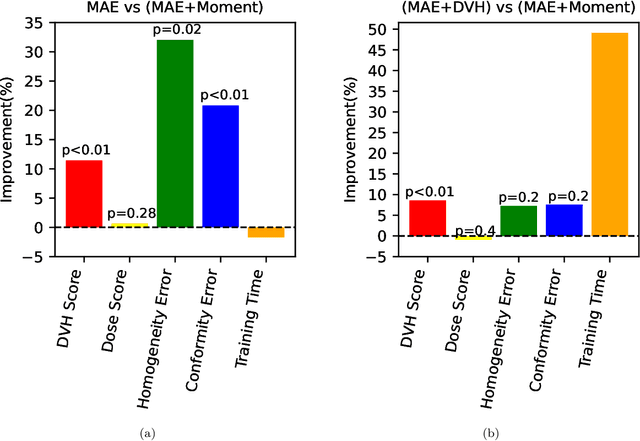
Abstract:Dose volume histogram (DVH) metrics are widely accepted evaluation criteria in the clinic. However, incorporating these metrics into deep learning dose prediction models is challenging due to their non-convexity and non-differentiability. We propose a novel moment-based loss function for predicting 3D dose distribution for the challenging conventional lung intensity modulated radiation therapy (IMRT) plans. The moment-based loss function is convex and differentiable and can easily incorporate DVH metrics in any deep learning framework without computational overhead. The moments can also be customized to reflect the clinical priorities in 3D dose prediction. For instance, using high-order moments allows better prediction in high-dose areas for serial structures. We used a large dataset of 360 (240 for training, 50 for validation and 70 for testing) conventional lung patients with 2Gy $\times$ 30 fractions to train the deep learning (DL) model using clinically treated plans at our institution. We trained a UNet like CNN architecture using computed tomography (CT), planning target volume (PTV) and organ-at-risk contours (OAR) as input to infer corresponding voxel-wise 3D dose distribution. We evaluated three different loss functions: (1) The popular Mean Absolute Error (MAE) Loss, (2) the recently developed MAE + DVH Loss, and (3) the proposed MAE + Moments Loss. The quality of the predictions was compared using different DVH metrics as well as dose-score and DVH-score, recently introduced by the AAPM knowledge-based planning grand challenge. Model with (MAE + Moment) loss function outperformed the model with MAE loss by significantly improving the DVH-score (11%, p$<$0.01) while having similar computational cost. It also outperformed the model trained with (MAE+DVH) by significantly improving the computational cost (48%) and the DVH-score (8%, p$<$0.01).
CIRDataset: A large-scale Dataset for Clinically-Interpretable lung nodule Radiomics and malignancy prediction
Jun 29, 2022



Abstract:Spiculations/lobulations, sharp/curved spikes on the surface of lung nodules, are good predictors of lung cancer malignancy and hence, are routinely assessed and reported by radiologists as part of the standardized Lung-RADS clinical scoring criteria. Given the 3D geometry of the nodule and 2D slice-by-slice assessment by radiologists, manual spiculation/lobulation annotation is a tedious task and thus no public datasets exist to date for probing the importance of these clinically-reported features in the SOTA malignancy prediction algorithms. As part of this paper, we release a large-scale Clinically-Interpretable Radiomics Dataset, CIRDataset, containing 956 radiologist QA/QC'ed spiculation/lobulation annotations on segmented lung nodules from two public datasets, LIDC-IDRI (N=883) and LUNGx (N=73). We also present an end-to-end deep learning model based on multi-class Voxel2Mesh extension to segment nodules (while preserving spikes), classify spikes (sharp/spiculation and curved/lobulation), and perform malignancy prediction. Previous methods have performed malignancy prediction for LIDC and LUNGx datasets but without robust attribution to any clinically reported/actionable features (due to known hyperparameter sensitivity issues with general attribution schemes). With the release of this comprehensively-annotated CIRDataset and end-to-end deep learning baseline, we hope that malignancy prediction methods can validate their explanations, benchmark against our baseline, and provide clinically-actionable insights. Dataset, code, pretrained models, and docker containers are available at https://github.com/nadeemlab/CIR.
Deep Learning 3D Dose Prediction for Conventional Lung IMRT Using Consistent/Unbiased Automated Plans
Jun 07, 2021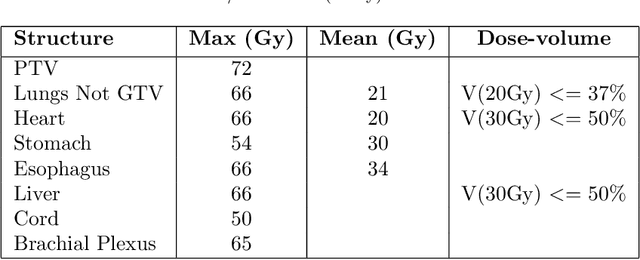
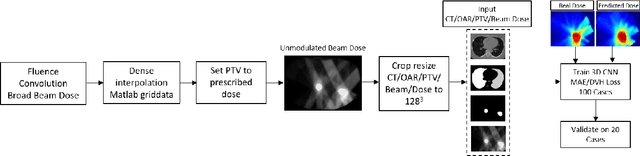
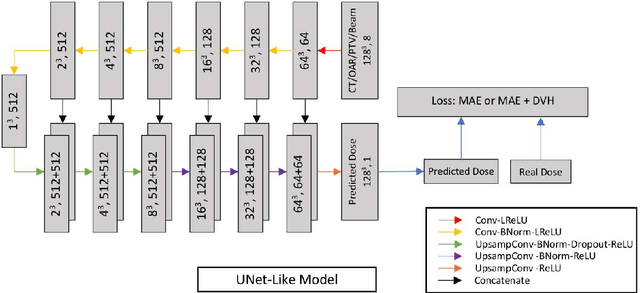

Abstract:Deep learning (DL) 3D dose prediction has recently gained a lot of attention. However, the variability of plan quality in the training dataset, generated manually by planners with wide range of expertise, can dramatically effect the quality of the final predictions. Moreover, any changes in the clinical criteria requires a new set of manually generated plans by planners to build a new prediction model. In this work, we instead use consistent plans generated by our in-house automated planning system (named ``ECHO'') to train the DL model. ECHO (expedited constrained hierarchical optimization) generates consistent/unbiased plans by solving large-scale constrained optimization problems sequentially. If the clinical criteria changes, a new training data set can be easily generated offline using ECHO, with no or limited human intervention, making the DL-based prediction model easily adaptable to the changes in the clinical practice. We used 120 conventional lung patients (100 for training, 20 for testing) with different beam configurations and trained our DL-model using manually-generated as well as automated ECHO plans. We evaluated different inputs: (1) CT+(PTV/OAR)contours, and (2) CT+contours+beam configurations, and different loss functions: (1) MAE (mean absolute error), and (2) MAE+DVH (dose volume histograms). The quality of the predictions was compared using different DVH metrics as well as dose-score and DVH-score, recently introduced by the AAPM knowledge-based planning grand challenge. The best results were obtained using automated ECHO plans and CT+contours+beam as training inputs and MAE+DVH as loss function.
An Efficiently Coupled Shape and Appearance Prior for Active Contour Segmentation
Mar 31, 2021
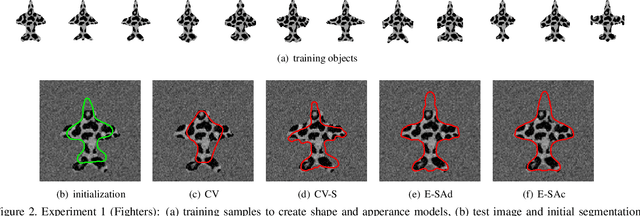
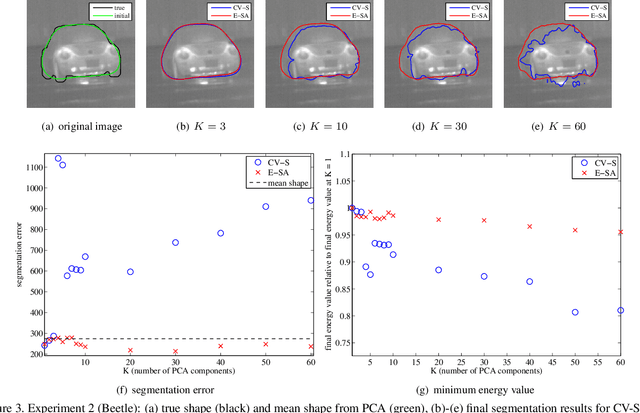
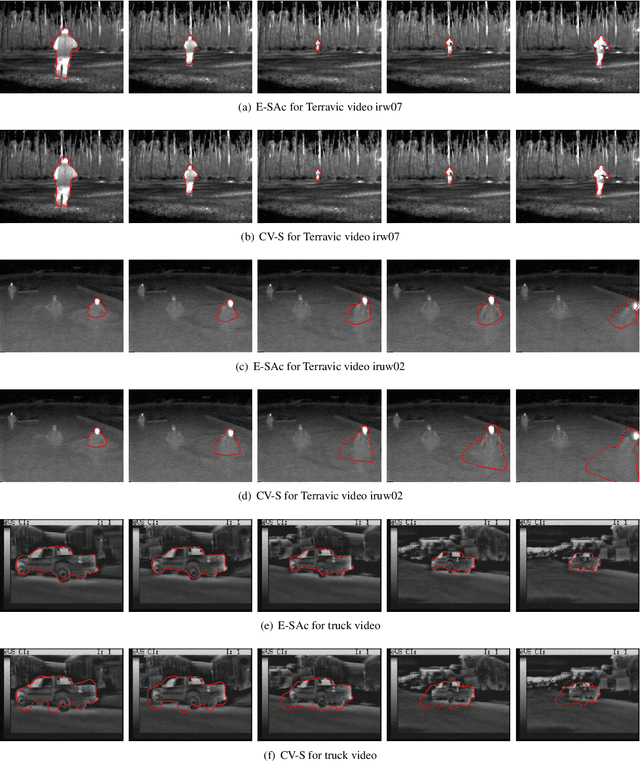
Abstract:This paper proposes a novel training model based on shape and appearance features for object segmentation in images and videos. Whereas most such models rely on two-dimensional appearance templates or a finite set of descriptors, our appearance-based feature is a one-dimensional function, which is efficiently coupled with the object's shape by integrating intensities along the object's iso-contours. Joint PCA training on these shape and appearance features further exploits shape-appearance correlations and the resulting training model is incorporated in an active-contour-type energy functional for recognition-segmentation tasks. Experiments on synthetic and infrared images demonstrate how this shape and appearance training model improves accuracy compared to methods based on the Chan-Vese energy.
Multitask 3D CBCT-to-CT Translation and Organs-at-Risk Segmentation Using Physics-Based Data Augmentation
Mar 09, 2021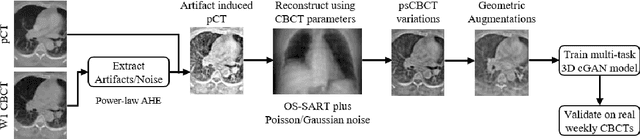
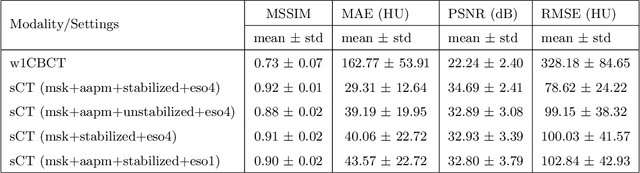
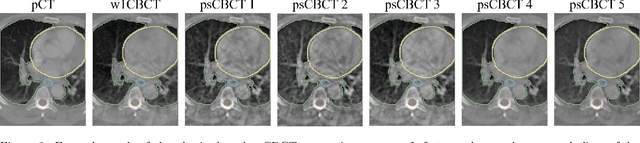
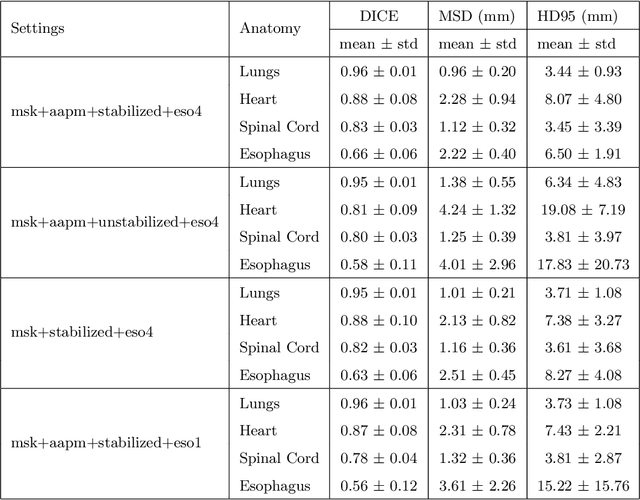
Abstract:Purpose: In current clinical practice, noisy and artifact-ridden weekly cone-beam computed tomography (CBCT) images are only used for patient setup during radiotherapy. Treatment planning is done once at the beginning of the treatment using high-quality planning CT (pCT) images and manual contours for organs-at-risk (OARs) structures. If the quality of the weekly CBCT images can be improved while simultaneously segmenting OAR structures, this can provide critical information for adapting radiotherapy mid-treatment as well as for deriving biomarkers for treatment response. Methods: Using a novel physics-based data augmentation strategy, we synthesize a large dataset of perfectly/inherently registered planning CT and synthetic-CBCT pairs for locally advanced lung cancer patient cohort, which are then used in a multitask 3D deep learning framework to simultaneously segment and translate real weekly CBCT images to high-quality planning CT-like images. Results: We compared the synthetic CT and OAR segmentations generated by the model to real planning CT and manual OAR segmentations and showed promising results. The real week 1 (baseline) CBCT images which had an average MAE of 162.77 HU compared to pCT images are translated to synthetic CT images that exhibit a drastically improved average MAE of 29.31 HU and average structural similarity of 92% with the pCT images. The average DICE scores of the 3D organs-at-risk segmentations are: lungs 0.96, heart 0.88, spinal cord 0.83 and esophagus 0.66. Conclusions: We demonstrate an approach to translate artifact-ridden CBCT images to high quality synthetic CT images while simultaneously generating good quality segmentation masks for different organs-at-risk. This approach could allow clinicians to adjust treatment plans using only the routine low-quality CBCT images, potentially improving patient outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge