Si-Yuan Zhang
Multitask 3D CBCT-to-CT Translation and Organs-at-Risk Segmentation Using Physics-Based Data Augmentation
Mar 09, 2021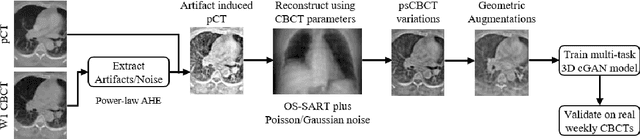
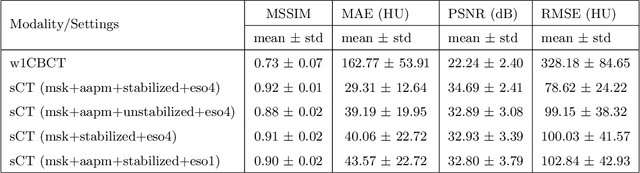
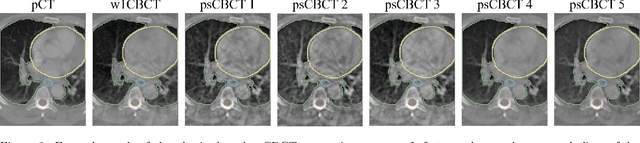
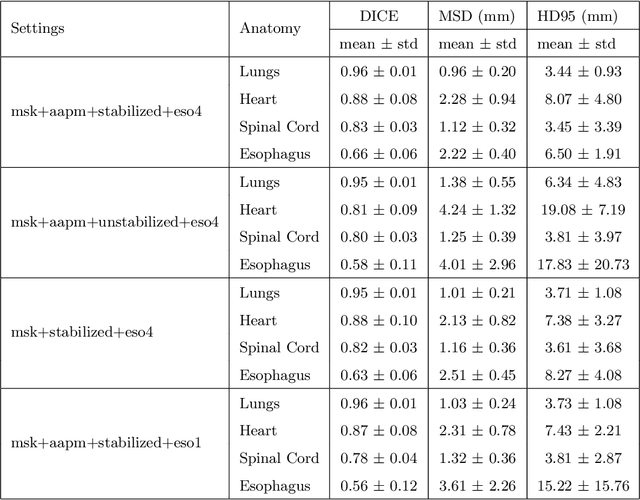
Abstract:Purpose: In current clinical practice, noisy and artifact-ridden weekly cone-beam computed tomography (CBCT) images are only used for patient setup during radiotherapy. Treatment planning is done once at the beginning of the treatment using high-quality planning CT (pCT) images and manual contours for organs-at-risk (OARs) structures. If the quality of the weekly CBCT images can be improved while simultaneously segmenting OAR structures, this can provide critical information for adapting radiotherapy mid-treatment as well as for deriving biomarkers for treatment response. Methods: Using a novel physics-based data augmentation strategy, we synthesize a large dataset of perfectly/inherently registered planning CT and synthetic-CBCT pairs for locally advanced lung cancer patient cohort, which are then used in a multitask 3D deep learning framework to simultaneously segment and translate real weekly CBCT images to high-quality planning CT-like images. Results: We compared the synthetic CT and OAR segmentations generated by the model to real planning CT and manual OAR segmentations and showed promising results. The real week 1 (baseline) CBCT images which had an average MAE of 162.77 HU compared to pCT images are translated to synthetic CT images that exhibit a drastically improved average MAE of 29.31 HU and average structural similarity of 92% with the pCT images. The average DICE scores of the 3D organs-at-risk segmentations are: lungs 0.96, heart 0.88, spinal cord 0.83 and esophagus 0.66. Conclusions: We demonstrate an approach to translate artifact-ridden CBCT images to high quality synthetic CT images while simultaneously generating good quality segmentation masks for different organs-at-risk. This approach could allow clinicians to adjust treatment plans using only the routine low-quality CBCT images, potentially improving patient outcomes.
Generalizable Cone Beam CT Esophagus Segmentation Using In Silico Data Augmentation
Jun 28, 2020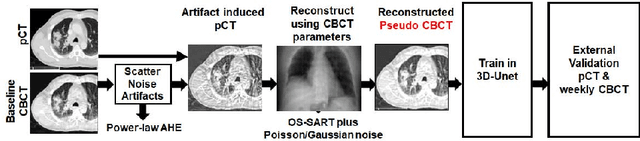
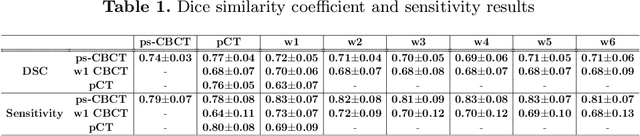
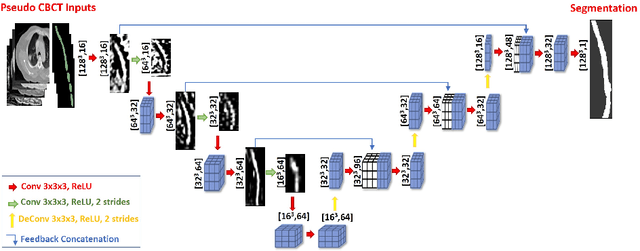

Abstract:Lung cancer radiotherapy entails high quality planning computed tomography (pCT) imaging of the patient with radiation oncologist contouring of the tumor and the organs at risk (OARs) at the start of the treatment. This is followed by weekly low-quality cone beam CT (CBCT) imaging for treatment setup and qualitative visual assessment of tumor and critical OARs. In this work, we aim to make the weekly CBCT assessment quantitative by automatically segmenting the most critical OAR, esophagus, using deep learning and in silico (image-driven simulation) artifact induction to convert pCTs to pseudo-CBCTs (pCTs$+$artifacts). Specifically, for the in silico data augmentation, we make use of the critical insight that CT and CBCT have the same underlying physics and that it is easier to deteriorate the pCT to look more like CBCT (and use the accompanying high quality manual contours for segmentation) than to synthesize CT from CBCT where the critical anatomical information may have already been lost (which leads to anatomical hallucination with the prevalent generative adversarial networks for example). Given these pseudo-CBCTs and the high quality manual contours, we introduce a modified 3D-Unet architecture and a multi-objective loss function specifically designed for segmenting soft-tissue organs such as esophagus on real weekly CBCTs. The model achieved 0.74 dice overlap (against manual contours of an experienced radiation oncologist) on weekly CBCTs and was robust and generalizable enough to also produce state-of-the-art results on pCTs, achieving 0.77 dice overlap against the previous best of 0.72. This shows that our in silico data augmentation spans the realistic noise/artifact spectrum across patient CBCT/pCT data and can generalize well across modalities (without requiring retraining or domain adaptation), eventually improving the accuracy of treatment setup and response analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge