Joseph Marino
Rethinking Histology Slide Digitization Workflows for Low-Resource Settings
May 13, 2024



Abstract:Histology slide digitization is becoming essential for telepathology (remote consultation), knowledge sharing (education), and using the state-of-the-art artificial intelligence algorithms (augmented/automated end-to-end clinical workflows). However, the cumulative costs of digital multi-slide high-speed brightfield scanners, cloud/on-premises storage, and personnel (IT and technicians) make the current slide digitization workflows out-of-reach for limited-resource settings, further widening the health equity gap; even single-slide manual scanning commercial solutions are costly due to hardware requirements (high-resolution cameras, high-spec PC/workstation, and support for only high-end microscopes). In this work, we present a new cloud slide digitization workflow for creating scanner-quality whole-slide images (WSIs) from uploaded low-quality videos, acquired from cheap and inexpensive microscopes with built-in cameras. Specifically, we present a pipeline to create stitched WSIs while automatically deblurring out-of-focus regions, upsampling input 10X images to 40X resolution, and reducing brightness/contrast and light-source illumination variations. We demonstrate the WSI creation efficacy from our workflow on World Health Organization-declared neglected tropical disease, Cutaneous Leishmaniasis (prevalent only in the poorest regions of the world and only diagnosed by sub-specialist dermatopathologists, rare in poor countries), as well as other common pathologies on core biopsies of breast, liver, duodenum, stomach and lymph node. The code and pretrained models will be accessible via our GitHub (https://github.com/nadeemlab/DeepLIIF), and the cloud platform will be available at https://deepliif.org for uploading microscope videos and downloading/viewing WSIs with shareable links (no sign-in required) for telepathology and knowledge sharing.
An AI-Ready Multiplex Staining Dataset for Reproducible and Accurate Characterization of Tumor Immune Microenvironment
May 25, 2023



Abstract:We introduce a new AI-ready computational pathology dataset containing restained and co-registered digitized images from eight head-and-neck squamous cell carcinoma patients. Specifically, the same tumor sections were stained with the expensive multiplex immunofluorescence (mIF) assay first and then restained with cheaper multiplex immunohistochemistry (mIHC). This is a first public dataset that demonstrates the equivalence of these two staining methods which in turn allows several use cases; due to the equivalence, our cheaper mIHC staining protocol can offset the need for expensive mIF staining/scanning which requires highly-skilled lab technicians. As opposed to subjective and error-prone immune cell annotations from individual pathologists (disagreement > 50%) to drive SOTA deep learning approaches, this dataset provides objective immune and tumor cell annotations via mIF/mIHC restaining for more reproducible and accurate characterization of tumor immune microenvironment (e.g. for immunotherapy). We demonstrate the effectiveness of this dataset in three use cases: (1) IHC quantification of CD3/CD8 tumor-infiltrating lymphocytes via style transfer, (2) virtual translation of cheap mIHC stains to more expensive mIF stains, and (3) virtual tumor/immune cellular phenotyping on standard hematoxylin images. The dataset is available at \url{https://github.com/nadeemlab/DeepLIIF}.
DeepLIIF: An Online Platform for Quantification of Clinical Pathology Slides
Apr 09, 2022



Abstract:In the clinic, resected tissue samples are stained with Hematoxylin-and-Eosin (H&E) and/or Immunhistochemistry (IHC) stains and presented to the pathologists on glass slides or as digital scans for diagnosis and assessment of disease progression. Cell-level quantification, e.g. in IHC protein expression scoring, can be extremely inefficient and subjective. We present DeepLIIF (https://deepliif.org), a first free online platform for efficient and reproducible IHC scoring. DeepLIIF outperforms current state-of-the-art approaches (relying on manual error-prone annotations) by virtually restaining clinical IHC slides with more informative multiplex immunofluorescence staining. Our DeepLIIF cloud-native platform supports (1) more than 150 proprietary/non-proprietary input formats via the Bio-Formats standard, (2) interactive adjustment, visualization, and downloading of the IHC quantification results and the accompanying restained images, (3) consumption of an exposed workflow API programmatically or through interactive plugins for open source whole slide image viewers such as QuPath/ImageJ, and (4) auto scaling to efficiently scale GPU resources based on user demand.
Insights from Generative Modeling for Neural Video Compression
Jul 28, 2021



Abstract:While recent machine learning research has revealed connections between deep generative models such as VAEs and rate-distortion losses used in learned compression, most of this work has focused on images. In a similar spirit, we view recently proposed neural video coding algorithms through the lens of deep autoregressive and latent variable modeling. We present recent neural video codecs as instances of a generalized stochastic temporal autoregressive transform, and propose new avenues for further improvements inspired by normalizing flows and structured priors. We propose several architectures that yield state-of-the-art video compression performance on full-resolution video and discuss their tradeoffs and ablations. In particular, we propose (i) improved temporal autoregressive transforms, (ii) improved entropy models with structured and temporal dependencies, and (iii) variable bitrate versions of our algorithms. Since our improvements are compatible with a large class of existing models, we provide further evidence that the generative modeling viewpoint can advance the neural video coding field.
Beyond Target Networks: Improving Deep $Q$-learning with Functional Regularization
Jun 07, 2021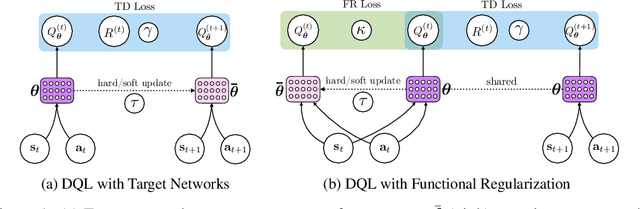
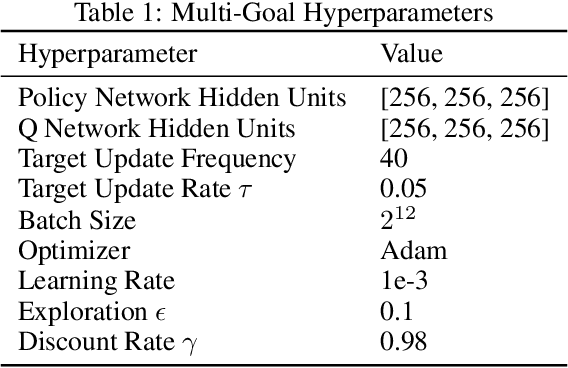
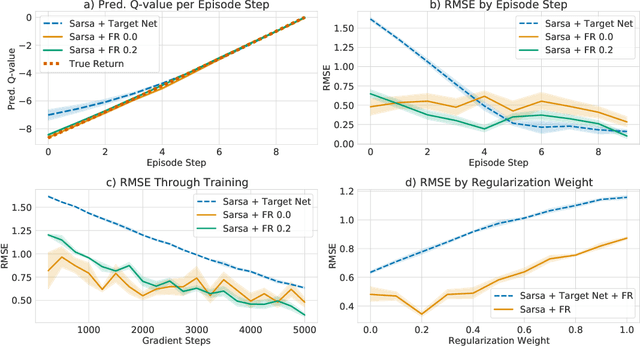
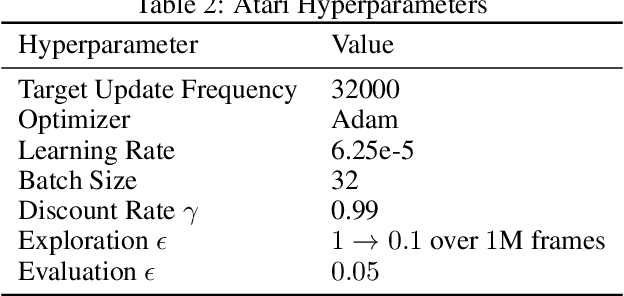
Abstract:Target networks are at the core of recent success in Reinforcement Learning. They stabilize the training by using old parameters to estimate the $Q$-values, but this also limits the propagation of newly-encountered rewards which could ultimately slow down the training. In this work, we propose an alternative training method based on functional regularization which does not have this deficiency. Unlike target networks, our method uses up-to-date parameters to estimate the target $Q$-values, thereby speeding up training while maintaining stability. Surprisingly, in some cases, we can show that target networks are a special, restricted type of functional regularizers. Using this approach, we show empirical improvements in sample efficiency and performance across a range of Atari and simulated robotics environments.
Predictive Coding, Variational Autoencoders, and Biological Connections
Nov 15, 2020

Abstract:This paper identifies connections between predictive coding, from theoretical neuroscience, and variational autoencoders, from machine learning. These connections imply striking correspondences for biological neural circuits, suggesting that pyramidal dendrites are functionally analogous to non-linear deep networks and lateral inhibition is functionally analogous to normalizing flows. Connecting these areas provides new directions for further investigations.
Iterative Amortized Policy Optimization
Oct 20, 2020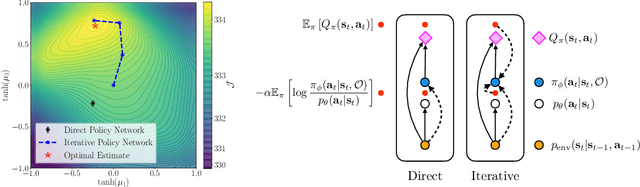
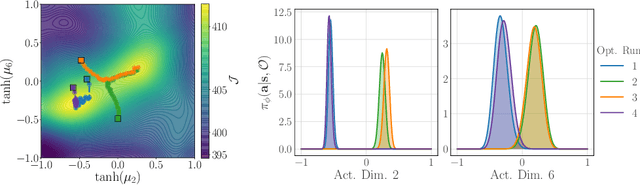
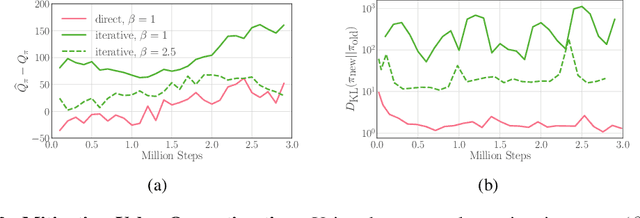
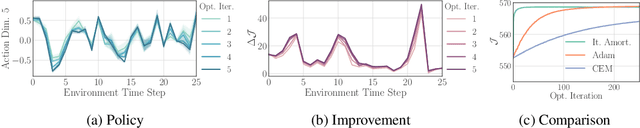
Abstract:Policy networks are a central feature of deep reinforcement learning (RL) algorithms for continuous control, enabling the estimation and sampling of high-value actions. From the variational inference perspective on RL, policy networks, when employed with entropy or KL regularization, are a form of amortized optimization, optimizing network parameters rather than the policy distributions directly. However, this direct amortized mapping can empirically yield suboptimal policy estimates. Given this perspective, we consider the more flexible class of iterative amortized optimizers. We demonstrate that the resulting technique, iterative amortized policy optimization, yields performance improvements over conventional direct amortization methods on benchmark continuous control tasks.
Hierarchical Autoregressive Modeling for Neural Video Compression
Oct 19, 2020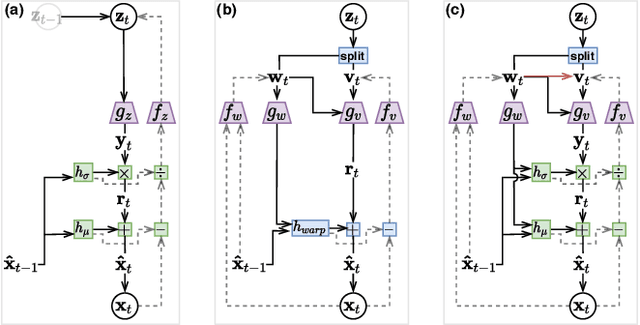
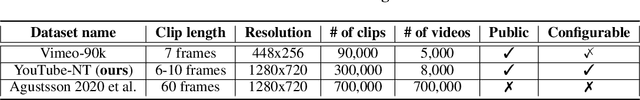
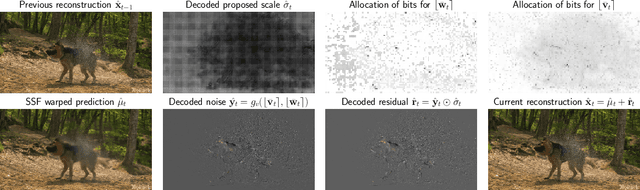
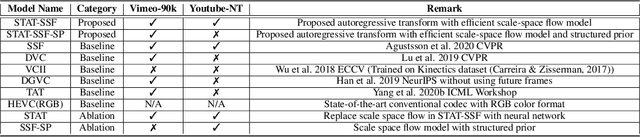
Abstract:Recent work by Marino et al. (2020) showed improved performance in sequential density estimation by combining masked autoregressive flows with hierarchical latent variable models. We draw a connection between such autoregressive generative models and the task of lossy video compression. Specifically, we view recent neural video compression methods (Lu et al., 2019; Yang et al., 2020b; Agustssonet al., 2020) as instances of a generalized stochastic temporal autoregressive trans-form, and propose avenues for enhancement based on this insight. Comprehensive evaluations on large-scale video data show improved rate-distortion performance over both state-of-the-art neural and conventional video compression methods.
Improving Sequential Latent Variable Models with Autoregressive Flows
Oct 07, 2020



Abstract:We propose an approach for improving sequence modeling based on autoregressive normalizing flows. Each autoregressive transform, acting across time, serves as a moving frame of reference, removing temporal correlations, and simplifying the modeling of higher-level dynamics. This technique provides a simple, general-purpose method for improving sequence modeling, with connections to existing and classical techniques. We demonstrate the proposed approach both with standalone flow-based models and as a component within sequential latent variable models. Results are presented on three benchmark video datasets, where autoregressive flow-based dynamics improve log-likelihood performance over baseline models. Finally, we illustrate the decorrelation and improved generalization properties of using flow-based dynamics.
A General Method for Amortizing Variational Filtering
Nov 13, 2018

Abstract:We introduce the variational filtering EM algorithm, a simple, general-purpose method for performing variational inference in dynamical latent variable models using information from only past and present variables, i.e. filtering. The algorithm is derived from the variational objective in the filtering setting and consists of an optimization procedure at each time step. By performing each inference optimization procedure with an iterative amortized inference model, we obtain a computationally efficient implementation of the algorithm, which we call amortized variational filtering. We present experiments demonstrating that this general-purpose method improves performance across several deep dynamical latent variable models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge