Anne Grabenstetter
Deep Multi-Magnification Networks for Multi-Class Breast Cancer Image Segmentation
Oct 29, 2019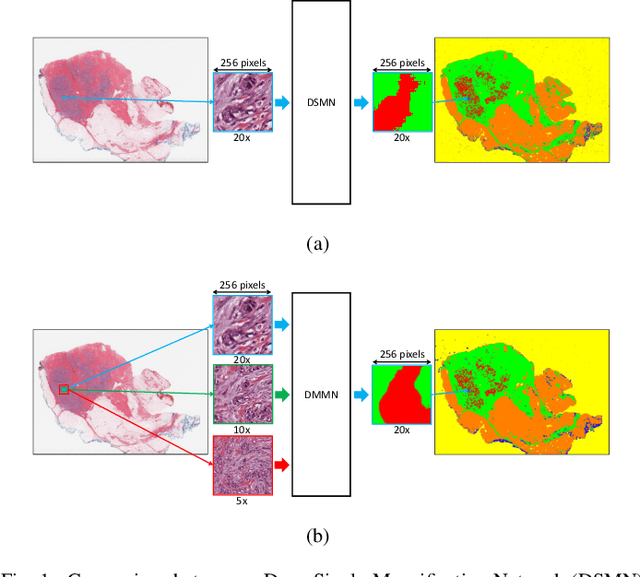
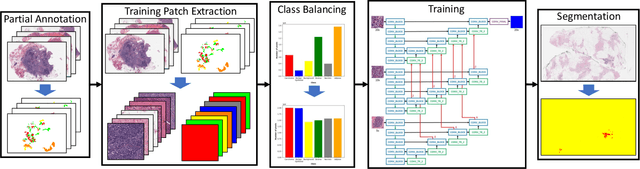
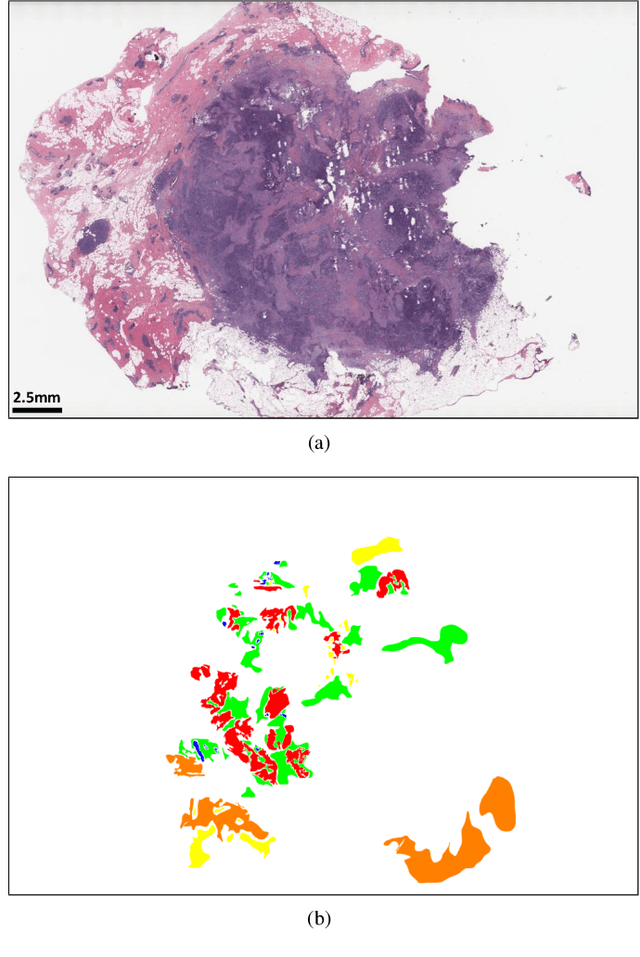
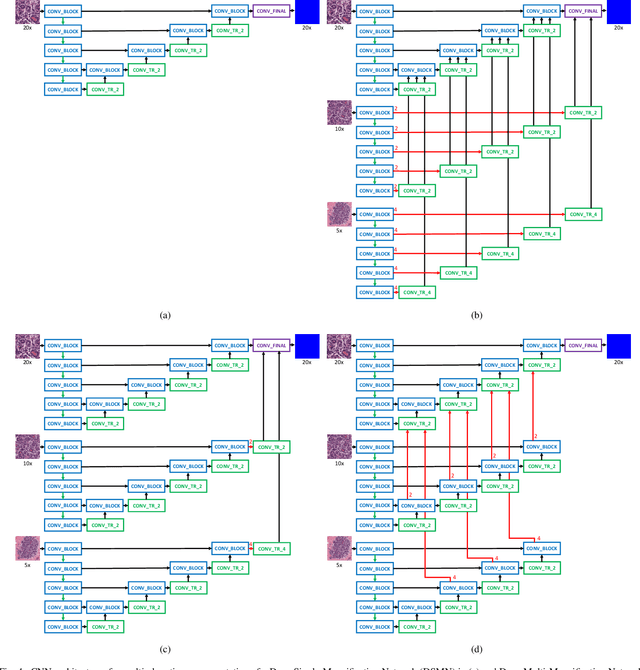
Abstract:Breast carcinoma is one of the most common cancers for women in the United States. Pathologic analysis of surgical excision specimens for breast carcinoma is important to evaluate the completeness of surgical excision and has implications for future treatment. This analysis is performed manually by pathologists reviewing histologic slides prepared from formalin-fixed tissue. Digital pathology has provided means to digitize the glass slides and generate whole slide images. Computational pathology enables whole slide images to be automatically analyzed to assist pathologists, especially with the advancement of deep learning. The whole slide images generally contain giga-pixels of data, so it is impractical to process the images at the whole-slide-level. Most of the current deep learning techniques process the images at the patch-level, but they may produce poor results by looking at individual patches with a narrow field-of-view at a single magnification. In this paper, we present Deep Multi-Magnification Networks (DMMNs) to resemble how pathologists analyze histologic slides using microscopes. Our multi-class tissue segmentation architecture processes a set of patches from multiple magnifications to make more accurate predictions. For our supervised training, we use partial annotations to reduce the burden of annotators. Our segmentation architecture with multi-encoder, multi-decoder, and multi-concatenation outperforms other segmentation architectures on breast datasets and can be used to facilitate pathologists' assessments of breast cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge