Thomas J. Fuchs
JPL
PRISM2: Unlocking Multi-Modal General Pathology AI with Clinical Dialogue
Jun 16, 2025Abstract:Recent pathology foundation models can provide rich tile-level representations but fall short of delivering general-purpose clinical utility without further extensive model development. These models lack whole-slide image (WSI) understanding and are not trained with large-scale diagnostic data, limiting their performance on diverse downstream tasks. We introduce PRISM2, a multi-modal slide-level foundation model trained via clinical dialogue to enable scalable, generalizable pathology AI. PRISM2 is trained on nearly 700,000 specimens (2.3 million WSIs) paired with real-world clinical diagnostic reports in a two-stage process. In Stage 1, a vision-language model is trained using contrastive and captioning objectives to align whole slide embeddings with textual clinical diagnosis. In Stage 2, the language model is unfrozen to enable diagnostic conversation and extract more clinically meaningful representations from hidden states. PRISM2 achieves strong performance on diagnostic and biomarker prediction tasks, outperforming prior slide-level models including PRISM and TITAN. It also introduces a zero-shot yes/no classification approach that surpasses CLIP-style methods without prompt tuning or class enumeration. By aligning visual features with clinical reasoning, PRISM2 improves generalization on both data-rich and low-sample tasks, offering a scalable path forward for building general pathology AI agents capable of assisting diagnostic and prognostic decisions.
Benchmarking Embedding Aggregation Methods in Computational Pathology: A Clinical Data Perspective
Jul 10, 2024


Abstract:Recent advances in artificial intelligence (AI), in particular self-supervised learning of foundation models (FMs), are revolutionizing medical imaging and computational pathology (CPath). A constant challenge in the analysis of digital Whole Slide Images (WSIs) is the problem of aggregating tens of thousands of tile-level image embeddings to a slide-level representation. Due to the prevalent use of datasets created for genomic research, such as TCGA, for method development, the performance of these techniques on diagnostic slides from clinical practice has been inadequately explored. This study conducts a thorough benchmarking analysis of ten slide-level aggregation techniques across nine clinically relevant tasks, including diagnostic assessment, biomarker classification, and outcome prediction. The results yield following key insights: (1) Embeddings derived from domain-specific (histological images) FMs outperform those from generic ImageNet-based models across aggregation methods. (2) Spatial-aware aggregators enhance the performance significantly when using ImageNet pre-trained models but not when using FMs. (3) No single model excels in all tasks and spatially-aware models do not show general superiority as it would be expected. These findings underscore the need for more adaptable and universally applicable aggregation techniques, guiding future research towards tools that better meet the evolving needs of clinical-AI in pathology. The code used in this work is available at \url{https://github.com/fuchs-lab-public/CPath_SABenchmark}.
PRISM: A Multi-Modal Generative Foundation Model for Slide-Level Histopathology
May 16, 2024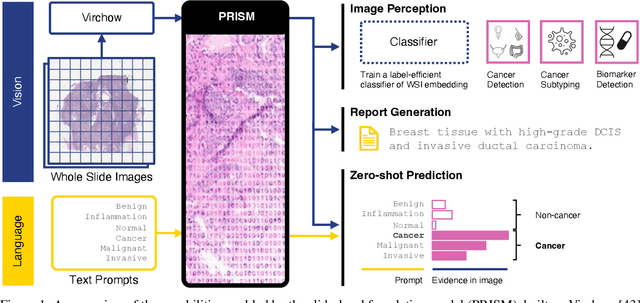
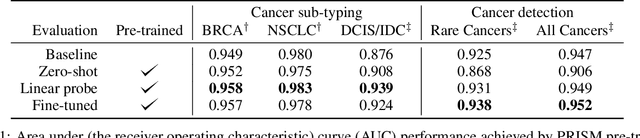
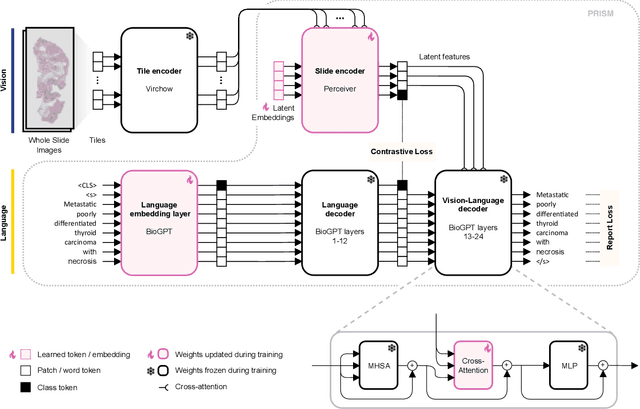
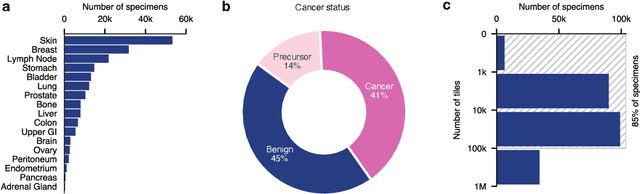
Abstract:Foundation models in computational pathology promise to unlock the development of new clinical decision support systems and models for precision medicine. However, there is a mismatch between most clinical analysis, which is defined at the level of one or more whole slide images, and foundation models to date, which process the thousands of image tiles contained in a whole slide image separately. The requirement to train a network to aggregate information across a large number of tiles in multiple whole slide images limits these models' impact. In this work, we present a slide-level foundation model for H&E-stained histopathology, PRISM, that builds on Virchow tile embeddings and leverages clinical report text for pre-training. Using the tile embeddings, PRISM produces slide-level embeddings with the ability to generate clinical reports, resulting in several modes of use. Using text prompts, PRISM achieves zero-shot cancer detection and sub-typing performance approaching and surpassing that of a supervised aggregator model. Using the slide embeddings with linear classifiers, PRISM surpasses supervised aggregator models. Furthermore, we demonstrate that fine-tuning of the PRISM slide encoder yields label-efficient training for biomarker prediction, a task that typically suffers from low availability of training data; an aggregator initialized with PRISM and trained on as little as 10% of the training data can outperform a supervised baseline that uses all of the data.
Beyond Multiple Instance Learning: Full Resolution All-In-Memory End-To-End Pathology Slide Modeling
Mar 07, 2024



Abstract:Artificial Intelligence (AI) has great potential to improve health outcomes by training systems on vast digitized clinical datasets. Computational Pathology, with its massive amounts of microscopy image data and impact on diagnostics and biomarkers, is at the forefront of this development. Gigapixel pathology slides pose a unique challenge due to their enormous size and are usually divided into tens of thousands of smaller tiles for analysis. This results in a discontinuity in the machine learning process by separating the training of tile-level encoders from slide-level aggregators and the need to adopt weakly supervised learning strategies. Training models from entire pathology slides end-to-end has been largely unexplored due to its computational challenges. To overcome this problem, we propose a novel approach to jointly train both a tile encoder and a slide-aggregator fully in memory and end-to-end at high-resolution, bridging the gap between input and slide-level supervision. While more computationally expensive, detailed quantitative validation shows promise for large-scale pre-training of pathology foundation models.
Computational Pathology at Health System Scale -- Self-Supervised Foundation Models from Three Billion Images
Oct 10, 2023Abstract:Recent breakthroughs in self-supervised learning have enabled the use of large unlabeled datasets to train visual foundation models that can generalize to a variety of downstream tasks. While this training paradigm is well suited for the medical domain where annotations are scarce, large-scale pre-training in the medical domain, and in particular pathology, has not been extensively studied. Previous work in self-supervised learning in pathology has leveraged smaller datasets for both pre-training and evaluating downstream performance. The aim of this project is to train the largest academic foundation model and benchmark the most prominent self-supervised learning algorithms by pre-training and evaluating downstream performance on large clinical pathology datasets. We collected the largest pathology dataset to date, consisting of over 3 billion images from over 423 thousand microscopy slides. We compared pre-training of visual transformer models using the masked autoencoder (MAE) and DINO algorithms. We evaluated performance on six clinically relevant tasks from three anatomic sites and two institutions: breast cancer detection, inflammatory bowel disease detection, breast cancer estrogen receptor prediction, lung adenocarcinoma EGFR mutation prediction, and lung cancer immunotherapy response prediction. Our results demonstrate that pre-training on pathology data is beneficial for downstream performance compared to pre-training on natural images. Additionally, the DINO algorithm achieved better generalization performance across all tasks tested. The presented results signify a phase change in computational pathology research, paving the way into a new era of more performant models based on large-scale, parallel pre-training at the billion-image scale.
Virchow: A Million-Slide Digital Pathology Foundation Model
Sep 21, 2023Abstract:Computational pathology uses artificial intelligence to enable precision medicine and decision support systems through the analysis of whole slide images. It has the potential to revolutionize the diagnosis and treatment of cancer. However, a major challenge to this objective is that for many specific computational pathology tasks the amount of data is inadequate for development. To address this challenge, we created Virchow, a 632 million parameter deep neural network foundation model for computational pathology. Using self-supervised learning, Virchow is trained on 1.5 million hematoxylin and eosin stained whole slide images from diverse tissue groups, which is orders of magnitude more data than previous works. When evaluated on downstream tasks including tile-level pan-cancer detection and subtyping and slide-level biomarker prediction, Virchow outperforms state-of-the-art systems both on internal datasets drawn from the same population as the pretraining data as well as external public datasets. Virchow achieves 93% balanced accuracy for pancancer tile classification, and AUCs of 0.983 for colon microsatellite instability status prediction and 0.967 for breast CDH1 status prediction. The gains in performance highlight the importance of pretraining on massive pathology image datasets, suggesting pretraining on even larger datasets could continue improving performance for many high-impact applications where limited amounts of training data are available, such as drug outcome prediction.
Deep conditional transformation models for survival analysis
Oct 20, 2022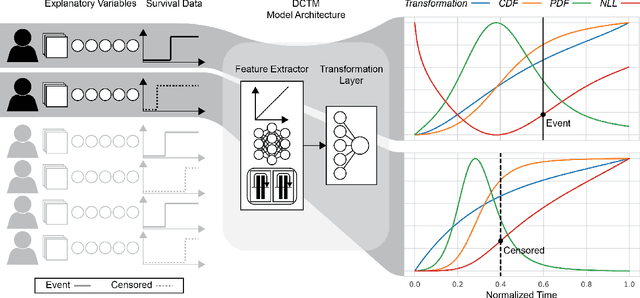
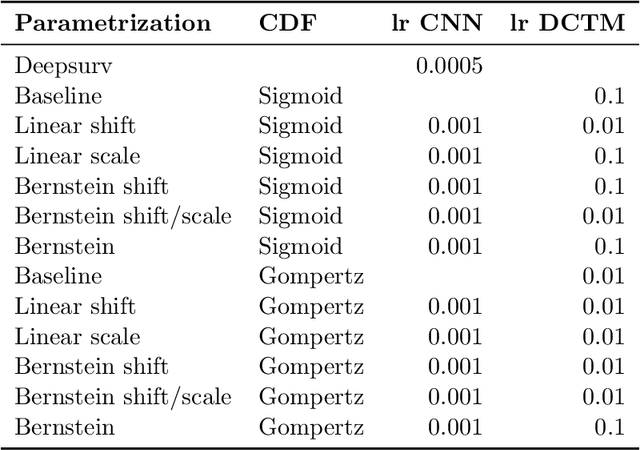
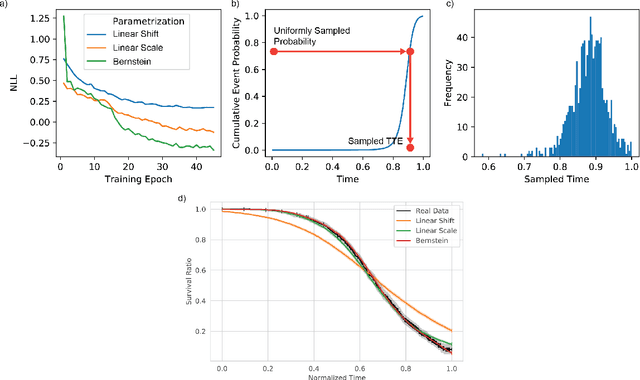
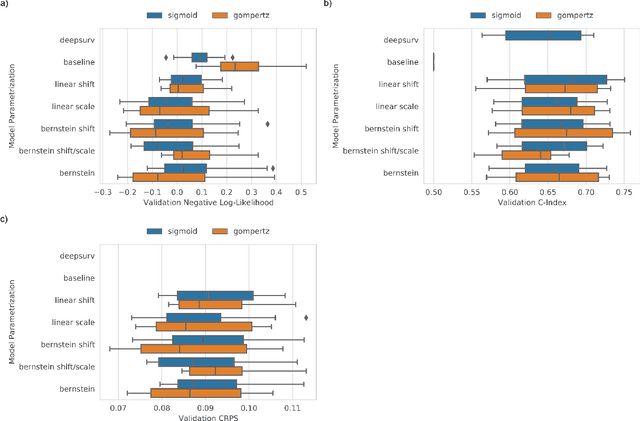
Abstract:An every increasing number of clinical trials features a time-to-event outcome and records non-tabular patient data, such as magnetic resonance imaging or text data in the form of electronic health records. Recently, several neural-network based solutions have been proposed, some of which are binary classifiers. Parametric, distribution-free approaches which make full use of survival time and censoring status have not received much attention. We present deep conditional transformation models (DCTMs) for survival outcomes as a unifying approach to parametric and semiparametric survival analysis. DCTMs allow the specification of non-linear and non-proportional hazards for both tabular and non-tabular data and extend to all types of censoring and truncation. On real and semi-synthetic data, we show that DCTMs compete with state-of-the-art DL approaches to survival analysis.
Deep Learning-Based Objective and Reproducible Osteosarcoma Chemotherapy Response Assessment and Outcome Prediction
Aug 09, 2022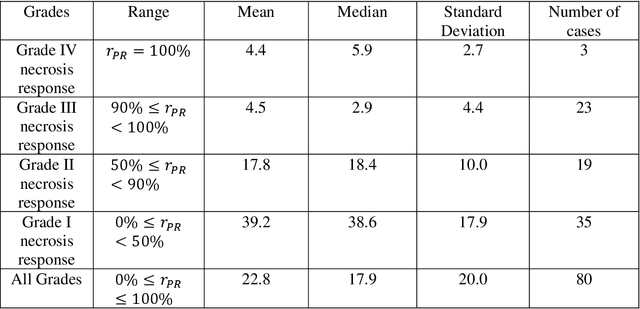
Abstract:Osteosarcoma is the most common primary bone cancer whose standard treatment includes pre-operative chemotherapy followed by resection. Chemotherapy response is used for predicting prognosis and further management of patients. Necrosis is routinely assessed post-chemotherapy from histology slides on resection specimens where necrosis ratio is defined as the ratio of necrotic tumor to overall tumor. Patients with necrosis ratio >=90% are known to have better outcome. Manual microscopic review of necrosis ratio from multiple glass slides is semi-quantitative and can have intra- and inter-observer variability. We propose an objective and reproducible deep learning-based approach to estimate necrosis ratio with outcome prediction from scanned hematoxylin and eosin whole slide images. We collected 103 osteosarcoma cases with 3134 WSIs to train our deep learning model, to validate necrosis ratio assessment, and to evaluate outcome prediction. We trained Deep Multi-Magnification Network to segment multiple tissue subtypes including viable tumor and necrotic tumor in pixel-level and to calculate case-level necrosis ratio from multiple WSIs. We showed necrosis ratio estimated by our segmentation model highly correlates with necrosis ratio from pathology reports manually assessed by experts where mean absolute differences for Grades IV (100%), III (>=90%), and II (>=50% and <90%) necrosis response are 4.4%, 4.5%, and 17.8%, respectively. We successfully stratified patients to predict overall survival with p=10^-6 and progression-free survival with p=0.012. Our reproducible approach without variability enabled us to tune cutoff thresholds, specifically for our model and our data set, to 80% for OS and 60% for PFS. Our study indicates deep learning can support pathologists as an objective tool to analyze osteosarcoma from histology for assessing treatment response and predicting patient outcome.
Deep Interactive Learning-based ovarian cancer segmentation of H&E-stained whole slide images to study morphological patterns of BRCA mutation
Mar 28, 2022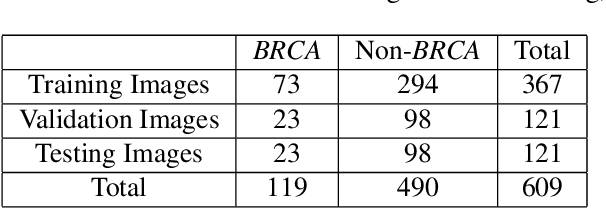
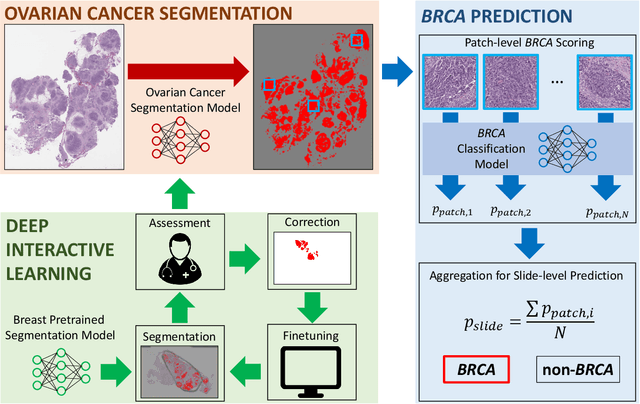
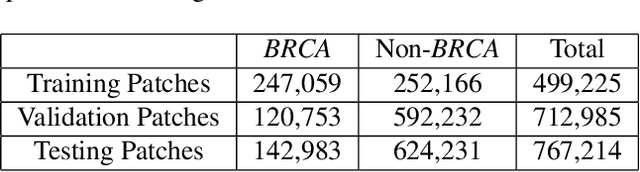
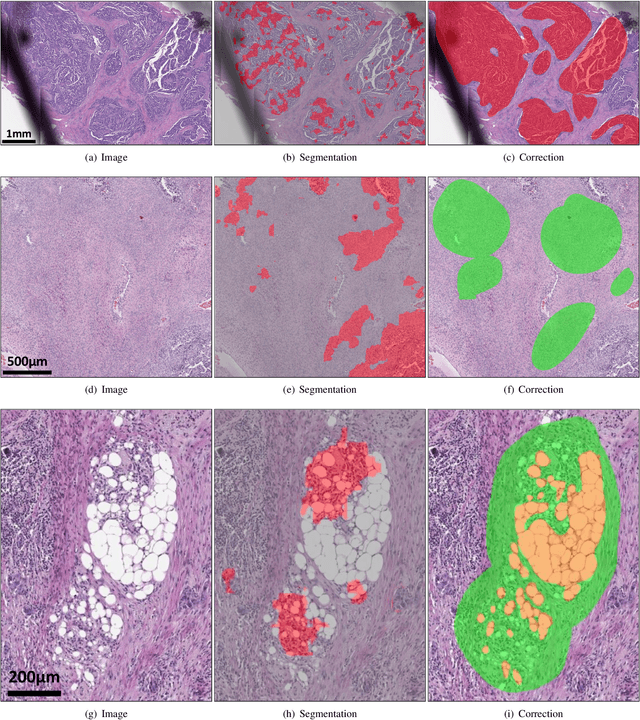
Abstract:Deep learning has been widely used to analyze digitized hematoxylin and eosin (H&E)-stained histopathology whole slide images. Automated cancer segmentation using deep learning can be used to diagnose malignancy and to find novel morphological patterns to predict molecular subtypes. To train pixel-wise cancer segmentation models, manual annotation from pathologists is generally a bottleneck due to its time-consuming nature. In this paper, we propose Deep Interactive Learning with a pretrained segmentation model from a different cancer type to reduce manual annotation time. Instead of annotating all pixels from cancer and non-cancer regions on giga-pixel whole slide images, an iterative process of annotating mislabeled regions from a segmentation model and training/finetuning the model with the additional annotation can reduce the time. Especially, employing a pretrained segmentation model can further reduce the time than starting annotation from scratch. We trained an accurate ovarian cancer segmentation model with a pretrained breast segmentation model by 3.5 hours of manual annotation which achieved intersection-over-union of 0.74, recall of 0.86, and precision of 0.84. With automatically extracted high-grade serous ovarian cancer patches, we attempted to train another deep learning model to predict BRCA mutation. The segmentation model and code have been released at https://github.com/MSKCC-Computational-Pathology/DMMN-ovary.
EPIC-Survival: End-to-end Part Inferred Clustering for Survival Analysis, Featuring Prognostic Stratification Boosting
Jan 28, 2021



Abstract:Histopathology-based survival modelling has two major hurdles. Firstly, a well-performing survival model has minimal clinical application if it does not contribute to the stratification of a cancer patient cohort into different risk groups, preferably driven by histologic morphologies. In the clinical setting, individuals are not given specific prognostic predictions, but are rather predicted to lie within a risk group which has a general survival trend. Thus, It is imperative that a survival model produces well-stratified risk groups. Secondly, until now, survival modelling was done in a two-stage approach (encoding and aggregation). The massive amount of pixels in digitized whole slide images were never utilized to their fullest extent due to technological constraints on data processing, forcing decoupled learning. EPIC-Survival bridges encoding and aggregation into an end-to-end survival modelling approach, while introducing stratification boosting to encourage the model to not only optimize ranking, but also to discriminate between risk groups. In this study we show that EPIC-Survival performs better than other approaches in modelling intrahepatic cholangiocarcinoma, a historically difficult cancer to model. Further, we show that stratification boosting improves further improves model performance, resulting in a concordance-index of 0.880 on a held-out test set. Finally, we were able to identify specific histologic differences, not commonly sought out in ICC, between low and high risk groups.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge