Eugene Fluder
Beyond Multiple Instance Learning: Full Resolution All-In-Memory End-To-End Pathology Slide Modeling
Mar 07, 2024



Abstract:Artificial Intelligence (AI) has great potential to improve health outcomes by training systems on vast digitized clinical datasets. Computational Pathology, with its massive amounts of microscopy image data and impact on diagnostics and biomarkers, is at the forefront of this development. Gigapixel pathology slides pose a unique challenge due to their enormous size and are usually divided into tens of thousands of smaller tiles for analysis. This results in a discontinuity in the machine learning process by separating the training of tile-level encoders from slide-level aggregators and the need to adopt weakly supervised learning strategies. Training models from entire pathology slides end-to-end has been largely unexplored due to its computational challenges. To overcome this problem, we propose a novel approach to jointly train both a tile encoder and a slide-aggregator fully in memory and end-to-end at high-resolution, bridging the gap between input and slide-level supervision. While more computationally expensive, detailed quantitative validation shows promise for large-scale pre-training of pathology foundation models.
Computational Pathology at Health System Scale -- Self-Supervised Foundation Models from Three Billion Images
Oct 10, 2023Abstract:Recent breakthroughs in self-supervised learning have enabled the use of large unlabeled datasets to train visual foundation models that can generalize to a variety of downstream tasks. While this training paradigm is well suited for the medical domain where annotations are scarce, large-scale pre-training in the medical domain, and in particular pathology, has not been extensively studied. Previous work in self-supervised learning in pathology has leveraged smaller datasets for both pre-training and evaluating downstream performance. The aim of this project is to train the largest academic foundation model and benchmark the most prominent self-supervised learning algorithms by pre-training and evaluating downstream performance on large clinical pathology datasets. We collected the largest pathology dataset to date, consisting of over 3 billion images from over 423 thousand microscopy slides. We compared pre-training of visual transformer models using the masked autoencoder (MAE) and DINO algorithms. We evaluated performance on six clinically relevant tasks from three anatomic sites and two institutions: breast cancer detection, inflammatory bowel disease detection, breast cancer estrogen receptor prediction, lung adenocarcinoma EGFR mutation prediction, and lung cancer immunotherapy response prediction. Our results demonstrate that pre-training on pathology data is beneficial for downstream performance compared to pre-training on natural images. Additionally, the DINO algorithm achieved better generalization performance across all tasks tested. The presented results signify a phase change in computational pathology research, paving the way into a new era of more performant models based on large-scale, parallel pre-training at the billion-image scale.
End-to-end Training for Whole Image Breast Cancer Screening using An All Convolutional Design
Sep 22, 2018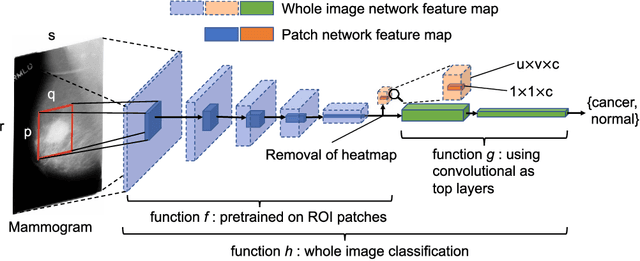



Abstract:An end-to-end training algorithm for the detection and classification of breast cancer on digital mammograms was created. In the initial training stage lesion annotations were used, but in subsequent stages, a whole image classifier was trained using only image-level labels, eliminating the reliance on rarely available lesion annotations. The simple all convolutional design provided superior performance in comparison with previous methods. For example, on the Digital Database for Screening Mammography (DDSM), the best single model achieved a per-image AUC of 0.88 on a holdout test set, and three-model averaging increased the AUC to 0.91. On an independent holdout set of images from the INbreast database, the best single model achieved a per-image AUC of 0.96. We also demonstrated that a whole image model trained on DDSM can be transferred to INbreast without using its lesion annotations and using only a small amount of INbreast data for fine-tuning. Code and model available at: https://github.com/lishen/end2end-all-conv
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge