Alican Bozkurt
Virchow: A Million-Slide Digital Pathology Foundation Model
Sep 21, 2023Abstract:Computational pathology uses artificial intelligence to enable precision medicine and decision support systems through the analysis of whole slide images. It has the potential to revolutionize the diagnosis and treatment of cancer. However, a major challenge to this objective is that for many specific computational pathology tasks the amount of data is inadequate for development. To address this challenge, we created Virchow, a 632 million parameter deep neural network foundation model for computational pathology. Using self-supervised learning, Virchow is trained on 1.5 million hematoxylin and eosin stained whole slide images from diverse tissue groups, which is orders of magnitude more data than previous works. When evaluated on downstream tasks including tile-level pan-cancer detection and subtyping and slide-level biomarker prediction, Virchow outperforms state-of-the-art systems both on internal datasets drawn from the same population as the pretraining data as well as external public datasets. Virchow achieves 93% balanced accuracy for pancancer tile classification, and AUCs of 0.983 for colon microsatellite instability status prediction and 0.967 for breast CDH1 status prediction. The gains in performance highlight the importance of pretraining on massive pathology image datasets, suggesting pretraining on even larger datasets could continue improving performance for many high-impact applications where limited amounts of training data are available, such as drug outcome prediction.
Segmentation of Cellular Patterns in Confocal Images of Melanocytic Lesions in vivo via a Multiscale Encoder-Decoder Network (MED-Net)
Jan 03, 2020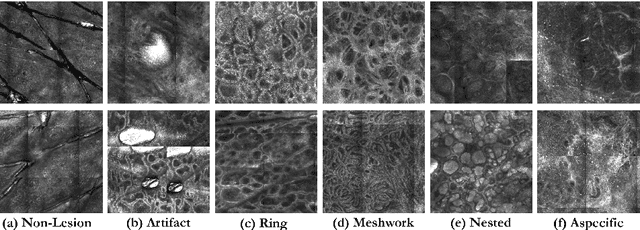
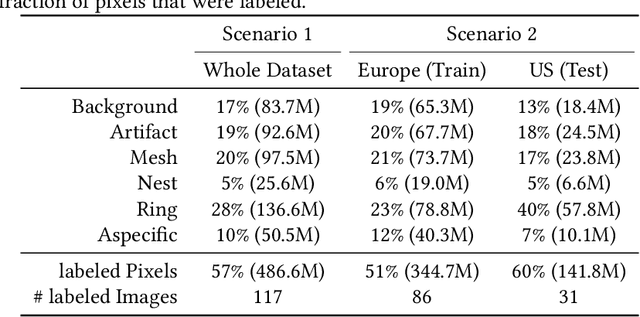
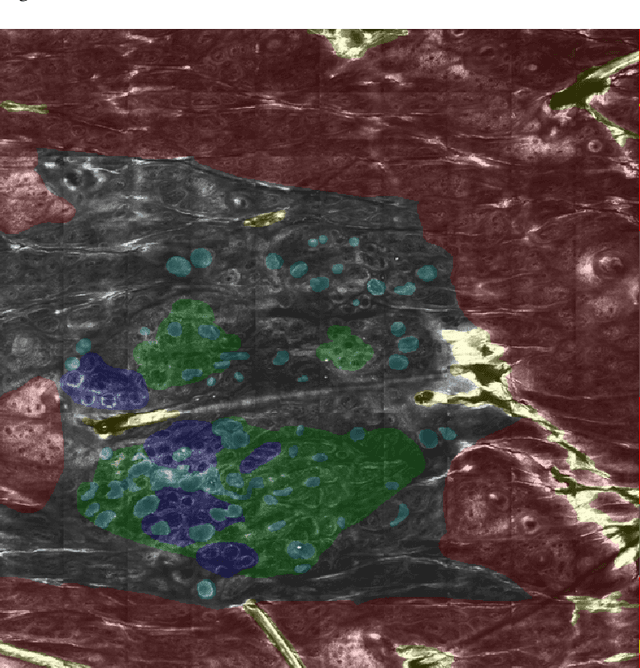
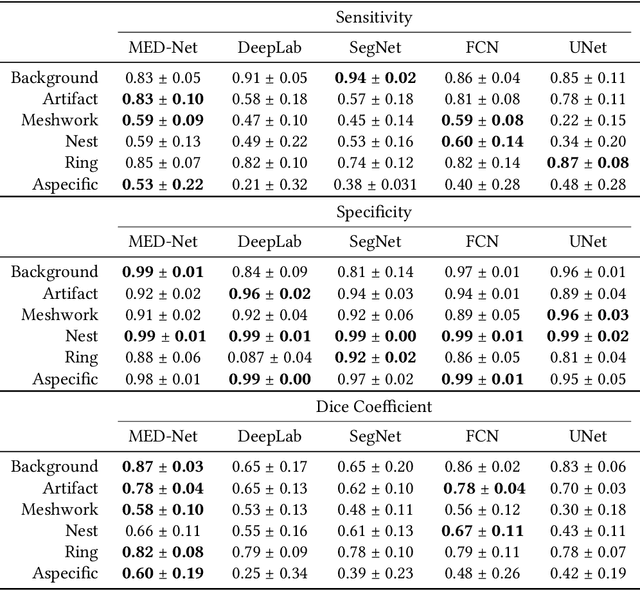
Abstract:In-vivo optical microscopy is advancing into routine clinical practice for non-invasively guiding diagnosis and treatment of cancer and other diseases, and thus beginning to reduce the need for traditional biopsy. However, reading and analysis of the optical microscopic images are generally still qualitative, relying mainly on visual examination. Here we present an automated semantic segmentation method called "Multiscale Encoder-Decoder Network (MED-Net)" that provides pixel-wise labeling into classes of patterns in a quantitative manner. The novelty in our approach is the modeling of textural patterns at multiple scales. This mimics the procedure for examining pathology images, which routinely starts with low magnification (low resolution, large field of view) followed by closer inspection of suspicious areas with higher magnification (higher resolution, smaller fields of view). We trained and tested our model on non-overlapping partitions of 117 reflectance confocal microscopy (RCM) mosaics of melanocytic lesions, an extensive dataset for this application, collected at four clinics in the US, and two in Italy. With patient-wise cross-validation, we achieved pixel-wise mean sensitivity and specificity of $70\pm11\%$ and $95\pm2\%$, respectively, with $0.71\pm0.09$ Dice coefficient over six classes. In the scenario, we partitioned the data clinic-wise and tested the generalizability of the model over multiple clinics. In this setting, we achieved pixel-wise mean sensitivity and specificity of $74\%$ and $95\%$, respectively, with $0.75$ Dice coefficient. We compared MED-Net against the state-of-the-art semantic segmentation models and achieved better quantitative segmentation performance. Our results also suggest that, due to its nested multiscale architecture, the MED-Net model annotated RCM mosaics more coherently, avoiding unrealistic-fragmented annotations.
Evaluating Combinatorial Generalization in Variational Autoencoders
Nov 11, 2019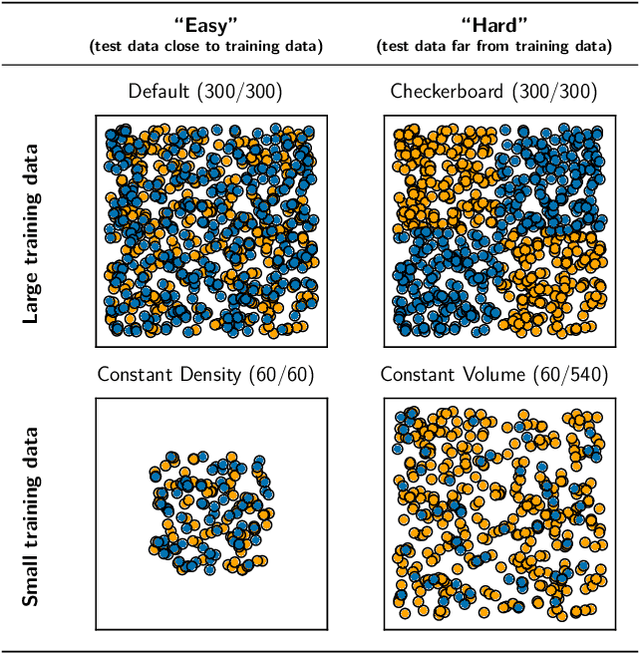
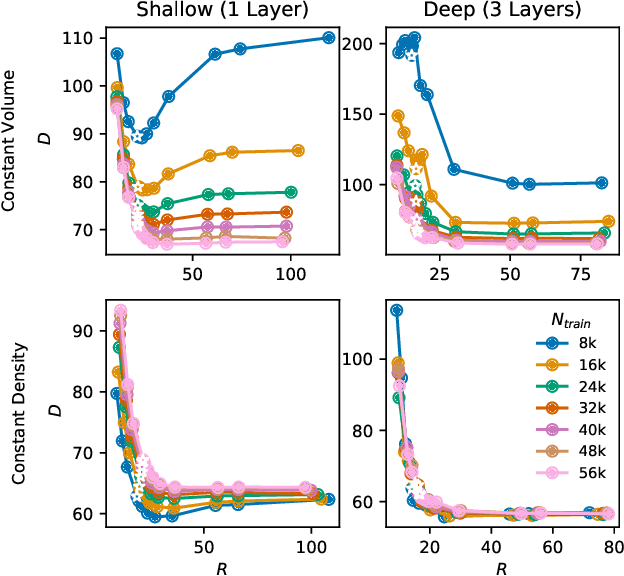
Abstract:We evaluate the ability of variational autoencoders to generalize to unseen examples in domains with a large combinatorial space of feature values. Our experiments systematically evaluate the effect of network width, depth, regularization, and the typical distance between the training and test examples. Increasing network capacity benefits generalization in easy problems, where test-set examples are similar to training examples. In more difficult problems, increasing capacity deteriorates generalization when optimizing the standard VAE objective, but once again improves generalization when we decrease the KL regularization. Our results establish that interplay between model capacity and KL regularization is not clear cut; we need to take the typical distance between train and test examples into account when evaluating generalization.
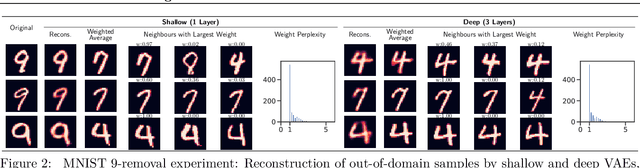
Can VAEs Generate Novel Examples?
Dec 22, 2018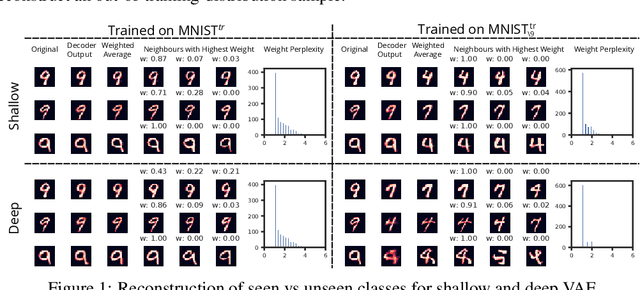
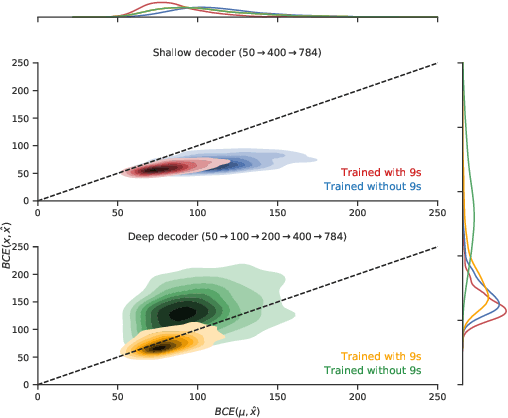

Abstract:An implicit goal in works on deep generative models is that such models should be able to generate novel examples that were not previously seen in the training data. In this paper, we investigate to what extent this property holds for widely employed variational autoencoder (VAE) architectures. VAEs maximize a lower bound on the log marginal likelihood, which implies that they will in principle overfit the training data when provided with a sufficiently expressive decoder. In the limit of an infinite capacity decoder, the optimal generative model is a uniform mixture over the training data. More generally, an optimal decoder should output a weighted average over the examples in the training data, where the magnitude of the weights is determined by the proximity in the latent space. This leads to the hypothesis that, for a sufficiently high capacity encoder and decoder, the VAE decoder will perform nearest-neighbor matching according to the coordinates in the latent space. To test this hypothesis, we investigate generalization on the MNIST dataset. We consider both generalization to new examples of previously seen classes, and generalization to the classes that were withheld from the training set. In both cases, we find that reconstructions are closely approximated by nearest neighbors for higher-dimensional parameterizations. When generalizing to unseen classes however, lower-dimensional parameterizations offer a clear advantage.
A Multiresolution Convolutional Neural Network with Partial Label Training for Annotating Reflectance Confocal Microscopy Images of Skin
Aug 23, 2018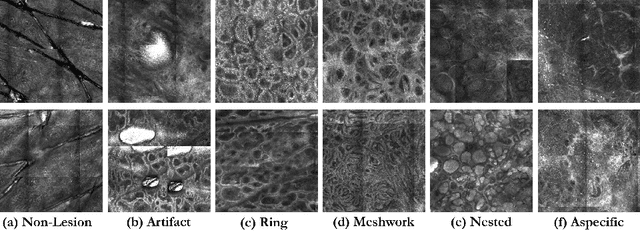
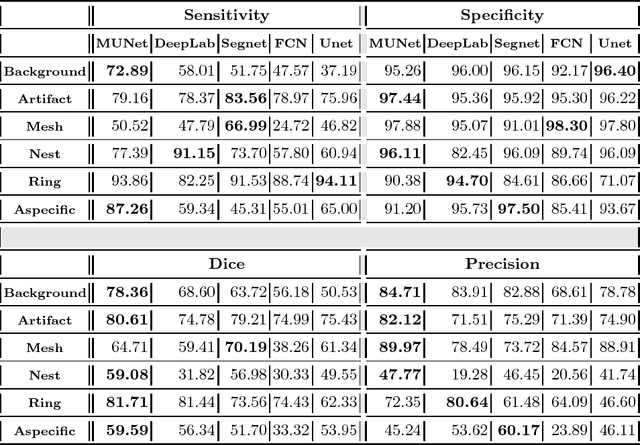
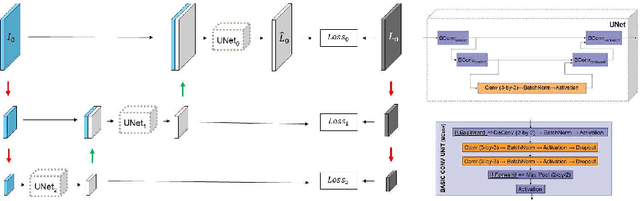
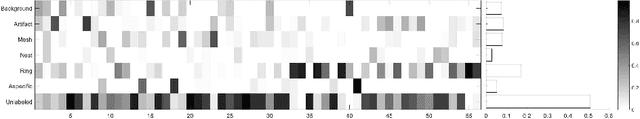
Abstract:We describe a new multiresolution "nested encoder-decoder" convolutional network architecture and use it to annotate morphological patterns in reflectance confocal microscopy (RCM) images of human skin for aiding cancer diagnosis. Skin cancers are the most common types of cancers, melanoma being the deadliest among them. RCM is an effective, non-invasive pre-screening tool for skin cancer diagnosis, with the required cellular resolution. However, images are complex, low-contrast, and highly variable, so that clinicians require months to years of expert-level training to be able to make accurate assessments. In this paper, we address classifying 4 key clinically important structural/textural patterns in RCM images. The occurrence and morphology of these patterns are used by clinicians for diagnosis of melanomas. The large size of RCM images, the large variance of pattern size, the large-scale range over which patterns appear, the class imbalance in collected images, and the lack of fully-labeled images all make this a challenging problem to address, even with automated machine learning tools. We designed a novel nested U-net architecture to cope with these challenges, and a selective loss function to handle partial labeling. Trained and tested on 56 melanoma-suspicious, partially labeled, 12k x 12k pixel images, our network automatically annotated diagnostic patterns with high sensitivity and specificity, providing consistent labels for unlabeled sections of the test images. Providing such annotation will aid clinicians in achieving diagnostic accuracy, and perhaps more important, dramatically facilitate clinical training, thus enabling much more rapid adoption of RCM into widespread clinical use process. In addition, our adaptation of U-net architecture provides an intrinsically multiresolution deep network that may be useful in other challenging biomedical image analysis applications.
Structured Disentangled Representations
May 29, 2018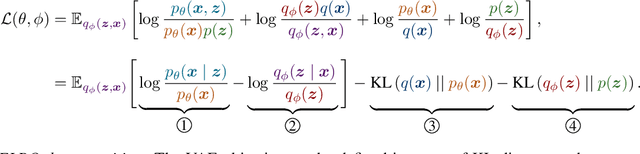
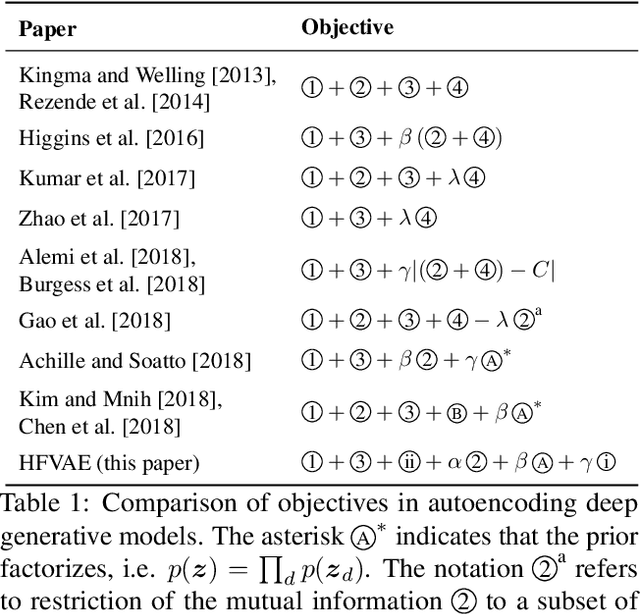
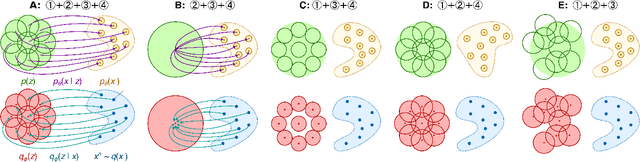
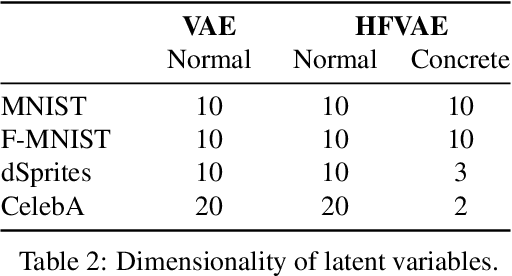
Abstract:Deep latent-variable models learn representations of high-dimensional data in an unsupervised manner. A number of recent efforts have focused on learning representations that disentangle statistically independent axes of variation by introducing modifications to the standard objective function. These approaches generally assume a simple diagonal Gaussian prior and as a result are not able to reliably disentangle discrete factors of variation. We propose a two-level hierarchical objective to control relative degree of statistical independence between blocks of variables and individual variables within blocks. We derive this objective as a generalization of the evidence lower bound, which allows us to explicitly represent the trade-offs between mutual information between data and representation, KL divergence between representation and prior, and coverage of the support of the empirical data distribution. Experiments on a variety of datasets demonstrate that our objective can not only disentangle discrete variables, but that doing so also improves disentanglement of other variables and, importantly, generalization even to unseen combinations of factors.
Delineation of Skin Strata in Reflectance Confocal Microscopy Images using Recurrent Convolutional Networks with Toeplitz Attention
Dec 01, 2017
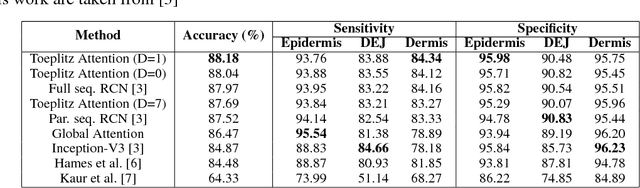
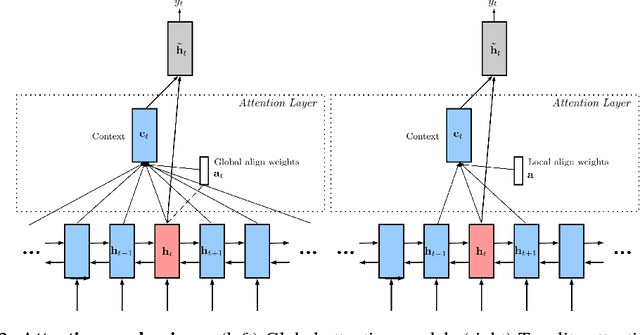
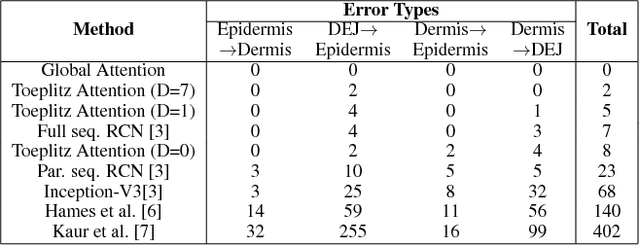
Abstract:Reflectance confocal microscopy (RCM) is an effective, non-invasive pre-screening tool for skin cancer diagnosis, but it requires extensive training and experience to assess accurately. There are few quantitative tools available to standardize image acquisition and analysis, and the ones that are available are not interpretable. In this study, we use a recurrent neural network with attention on convolutional network features. We apply it to delineate skin strata in vertically-oriented stacks of transverse RCM image slices in an interpretable manner. We introduce a new attention mechanism called Toeplitz attention, which constrains the attention map to have a Toeplitz structure. Testing our model on an expert labeled dataset of 504 RCM stacks, we achieve 88.17% image-wise classification accuracy, which is the current state-of-art.
Classifying Fonts and Calligraphy Styles Using Complex Wavelet Transform
Jul 09, 2014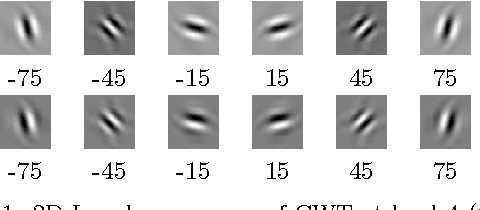

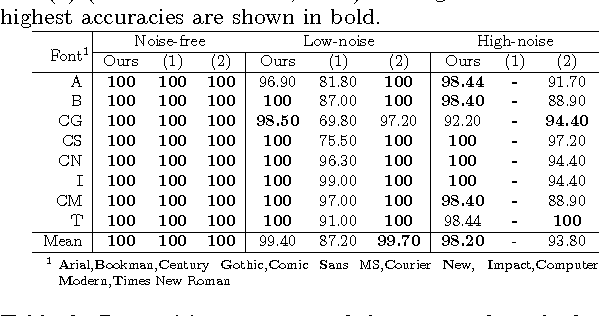
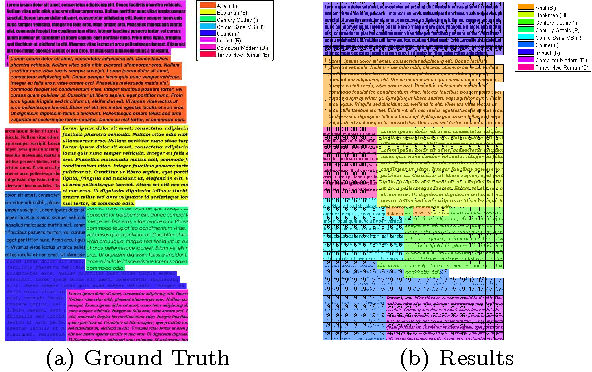
Abstract:Recognizing fonts has become an important task in document analysis, due to the increasing number of available digital documents in different fonts and emphases. A generic font-recognition system independent of language, script and content is desirable for processing various types of documents. At the same time, categorizing calligraphy styles in handwritten manuscripts is important for palaeographic analysis, but has not been studied sufficiently in the literature. We address the font-recognition problem as analysis and categorization of textures. We extract features using complex wavelet transform and use support vector machines for classification. Extensive experimental evaluations on different datasets in four languages and comparisons with state-of-the-art studies show that our proposed method achieves higher recognition accuracy while being computationally simpler. Furthermore, on a new dataset generated from Ottoman manuscripts, we show that the proposed method can also be used for categorizing Ottoman calligraphy with high accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge