Segmentation of Cellular Patterns in Confocal Images of Melanocytic Lesions in vivo via a Multiscale Encoder-Decoder Network (MED-Net)
Paper and Code
Jan 03, 2020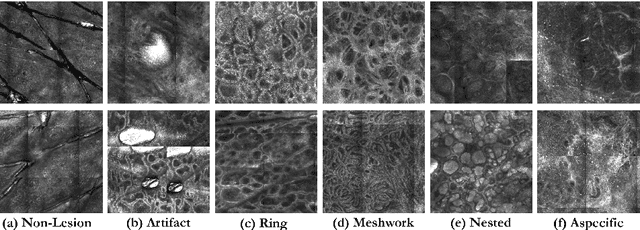
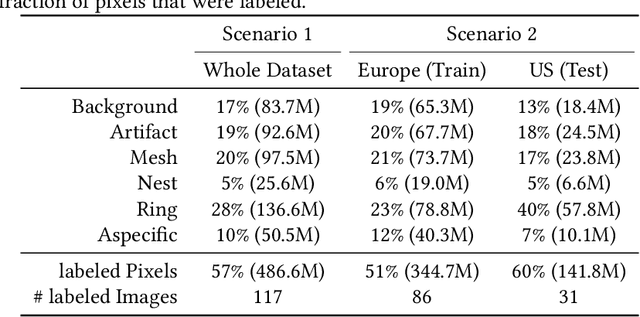
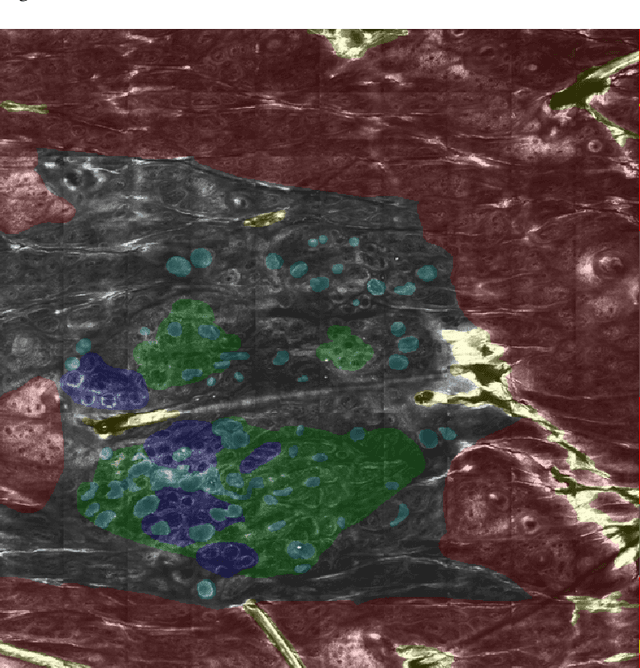
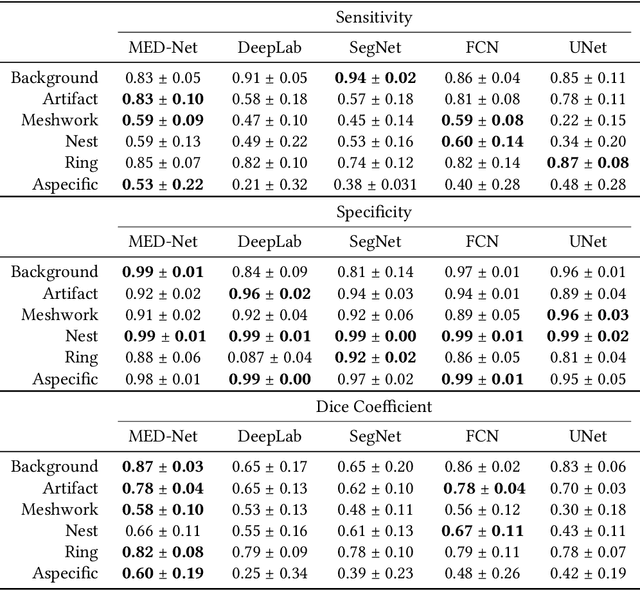
In-vivo optical microscopy is advancing into routine clinical practice for non-invasively guiding diagnosis and treatment of cancer and other diseases, and thus beginning to reduce the need for traditional biopsy. However, reading and analysis of the optical microscopic images are generally still qualitative, relying mainly on visual examination. Here we present an automated semantic segmentation method called "Multiscale Encoder-Decoder Network (MED-Net)" that provides pixel-wise labeling into classes of patterns in a quantitative manner. The novelty in our approach is the modeling of textural patterns at multiple scales. This mimics the procedure for examining pathology images, which routinely starts with low magnification (low resolution, large field of view) followed by closer inspection of suspicious areas with higher magnification (higher resolution, smaller fields of view). We trained and tested our model on non-overlapping partitions of 117 reflectance confocal microscopy (RCM) mosaics of melanocytic lesions, an extensive dataset for this application, collected at four clinics in the US, and two in Italy. With patient-wise cross-validation, we achieved pixel-wise mean sensitivity and specificity of $70\pm11\%$ and $95\pm2\%$, respectively, with $0.71\pm0.09$ Dice coefficient over six classes. In the scenario, we partitioned the data clinic-wise and tested the generalizability of the model over multiple clinics. In this setting, we achieved pixel-wise mean sensitivity and specificity of $74\%$ and $95\%$, respectively, with $0.75$ Dice coefficient. We compared MED-Net against the state-of-the-art semantic segmentation models and achieved better quantitative segmentation performance. Our results also suggest that, due to its nested multiscale architecture, the MED-Net model annotated RCM mosaics more coherently, avoiding unrealistic-fragmented annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge