Yubo Fan
IntraStyler: Exemplar-based Style Synthesis for Cross-modality Domain Adaptation
Jan 01, 2026Abstract:Image-level domain alignment is the de facto approach for unsupervised domain adaptation, where unpaired image translation is used to minimize the domain gap. Prior studies mainly focus on the domain shift between the source and target domains, whereas the intra-domain variability remains under-explored. To address the latter, an effective strategy is to diversify the styles of the synthetic target domain data during image translation. However, previous methods typically require intra-domain variations to be pre-specified for style synthesis, which may be impractical. In this paper, we propose an exemplar-based style synthesis method named IntraStyler, which can capture diverse intra-domain styles without any prior knowledge. Specifically, IntraStyler uses an exemplar image to guide the style synthesis such that the output style matches the exemplar style. To extract the style-only features, we introduce a style encoder to learn styles discriminatively based on contrastive learning. We evaluate the proposed method on the largest public dataset for cross-modality domain adaptation, CrossMoDA 2023. Our experiments show the efficacy of our method in controllable style synthesis and the benefits of diverse synthetic data for downstream segmentation. Code is available at https://github.com/han-liu/IntraStyler.
Human Locomotion Implicit Modeling Based Real-Time Gait Phase Estimation
Jun 18, 2025Abstract:Gait phase estimation based on inertial measurement unit (IMU) signals facilitates precise adaptation of exoskeletons to individual gait variations. However, challenges remain in achieving high accuracy and robustness, particularly during periods of terrain changes. To address this, we develop a gait phase estimation neural network based on implicit modeling of human locomotion, which combines temporal convolution for feature extraction with transformer layers for multi-channel information fusion. A channel-wise masked reconstruction pre-training strategy is proposed, which first treats gait phase state vectors and IMU signals as joint observations of human locomotion, thus enhancing model generalization. Experimental results demonstrate that the proposed method outperforms existing baseline approaches, achieving a gait phase RMSE of $2.729 \pm 1.071%$ and phase rate MAE of $0.037 \pm 0.016%$ under stable terrain conditions with a look-back window of 2 seconds, and a phase RMSE of $3.215 \pm 1.303%$ and rate MAE of $0.050 \pm 0.023%$ under terrain transitions. Hardware validation on a hip exoskeleton further confirms that the algorithm can reliably identify gait cycles and key events, adapting to various continuous motion scenarios. This research paves the way for more intelligent and adaptive exoskeleton systems, enabling safer and more efficient human-robot interaction across diverse real-world environments.
Predicting fluorescent labels in label-free microscopy images with pix2pix and adaptive loss in Light My Cells challenge
Jun 22, 2024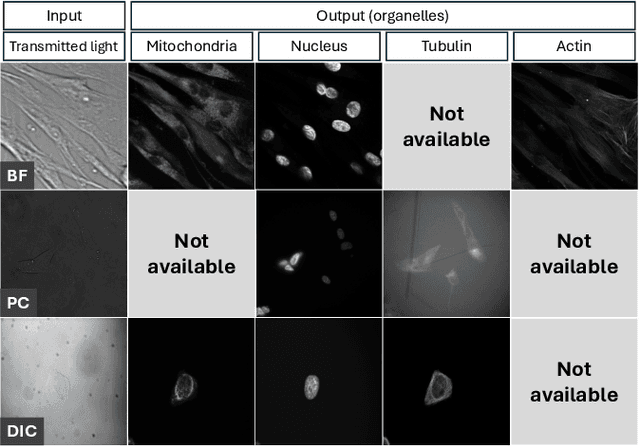
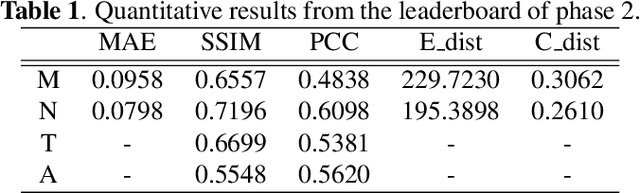
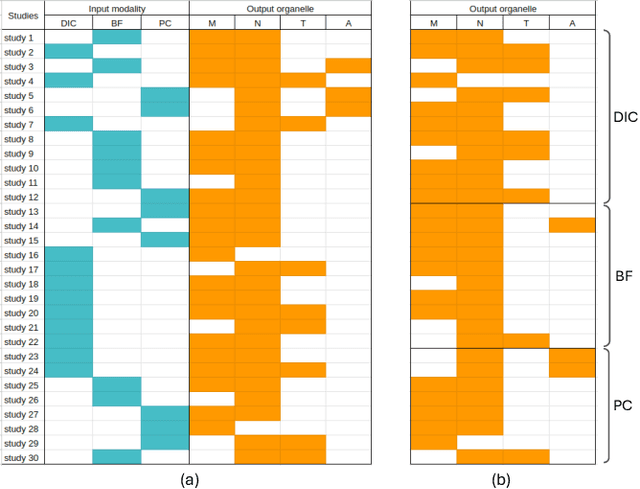
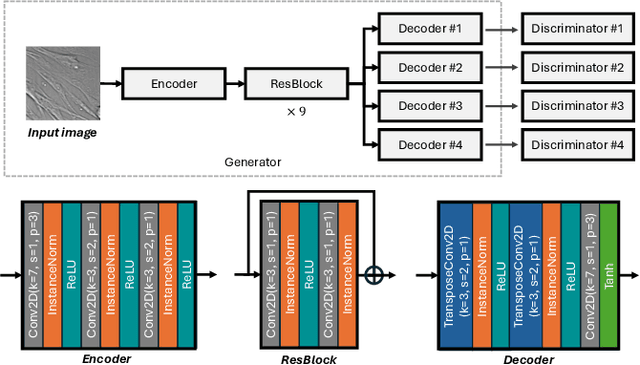
Abstract:Fluorescence labeling is the standard approach to reveal cellular structures and other subcellular constituents for microscopy images. However, this invasive procedure may perturb or even kill the cells and the procedure itself is highly time-consuming and complex. Recently, in silico labeling has emerged as a promising alternative, aiming to use machine learning models to directly predict the fluorescently labeled images from label-free microscopy. In this paper, we propose a deep learning-based in silico labeling method for the Light My Cells challenge. Built upon pix2pix, our proposed method can be trained using the partially labeled datasets with an adaptive loss. Moreover, we explore the effectiveness of several training strategies to handle different input modalities, such as training them together or separately. The results show that our method achieves promising performance for in silico labeling. Our code is available at https://github.com/MedICL-VU/LightMyCells.
Terrain-Aware Stride-Level Trajectory Forecasting for a Powered Hip Exoskeleton via Vision and Kinematics Fusion
Apr 18, 2024Abstract:Powered hip exoskeletons have shown the ability for locomotion assistance during treadmill walking. However, providing suitable assistance in real-world walking scenarios which involve changing terrain remains challenging. Recent research suggests that forecasting the lower limb joint's angles could provide target trajectories for exoskeletons and prostheses, and the performance could be improved with visual information. In this letter, We share a real-world dataset of 10 healthy subjects walking through five common types of terrain with stride-level label. We design a network called Sandwich Fusion Transformer for Image and Kinematics (SFTIK), which predicts the thigh angle of the ensuing stride given the terrain images at the beginning of the preceding and the ensuing stride and the IMU time series during the preceding stride. We introduce width-level patchify, tailored for egocentric terrain images, to reduce the computational demands. We demonstrate the proposed sandwich input and fusion mechanism could significantly improve the forecasting performance. Overall, the SFTIK outperforms baseline methods, achieving a computational efficiency of 3.31 G Flops, and root mean square error (RMSE) of 3.445 \textpm \ 0.804\textdegree \ and Pearson's correlation coefficient (PCC) of 0.971 \textpm\ 0.025. The results demonstrate that SFTIK could forecast the thigh's angle accurately with low computational cost, which could serve as a terrain adaptive trajectory planning method for hip exoskeletons. Codes and data are available at https://github.com/RuoqiZhao116/SFTIK.
Learning Site-specific Styles for Multi-institutional Unsupervised Cross-modality Domain Adaptation
Nov 22, 2023



Abstract:Unsupervised cross-modality domain adaptation is a challenging task in medical image analysis, and it becomes more challenging when source and target domain data are collected from multiple institutions. In this paper, we present our solution to tackle the multi-institutional unsupervised domain adaptation for the crossMoDA 2023 challenge. First, we perform unpaired image translation to translate the source domain images to the target domain, where we design a dynamic network to generate synthetic target domain images with controllable, site-specific styles. Afterwards, we train a segmentation model using the synthetic images and further reduce the domain gap by self-training. Our solution achieved the 1st place during both the validation and testing phases of the challenge. The code repository is publicly available at https://github.com/MedICL-VU/crossmoda2023.
MLP Based Continuous Gait Recognition of a Powered Ankle Prosthesis with Serial Elastic Actuator
Sep 15, 2023Abstract:Powered ankle prostheses effectively assist people with lower limb amputation to perform daily activities. High performance prostheses with adjustable compliance and capability to predict and implement amputee's intent are crucial for them to be comparable to or better than a real limb. However, current designs fail to provide simple yet effective compliance of the joint with full potential of modification, and lack accurate gait prediction method in real time. This paper proposes an innovative design of powered ankle prosthesis with serial elastic actuator (SEA), and puts forward a MLP based gait recognition method that can accurately and continuously predict more gait parameters for motion sensing and control. The prosthesis mimics biological joint with similar weight, torque, and power which can assist walking of up to 4 m/s. A new design of planar torsional spring is proposed for the SEA, which has better stiffness, endurance, and potential of modification than current designs. The gait recognition system simultaneously generates locomotive speed, gait phase, ankle angle and angular velocity only utilizing signals of single IMU, holding advantage in continuity, adaptability for speed range, accuracy, and capability of multi-functions.
Nucleus-aware Self-supervised Pretraining Using Unpaired Image-to-image Translation for Histopathology Images
Sep 14, 2023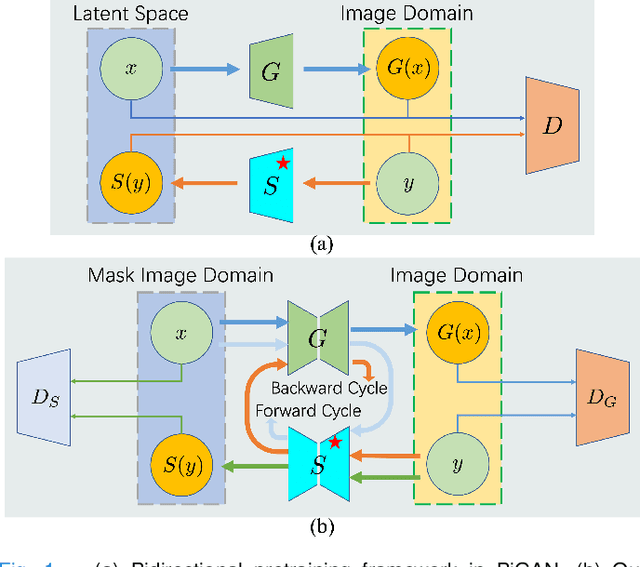
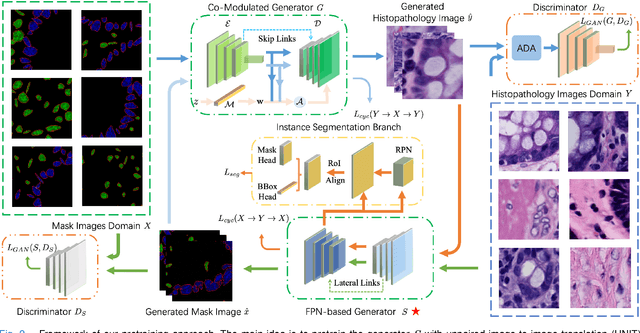
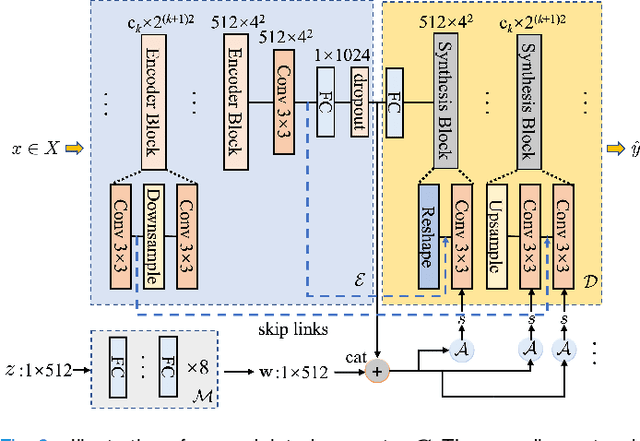
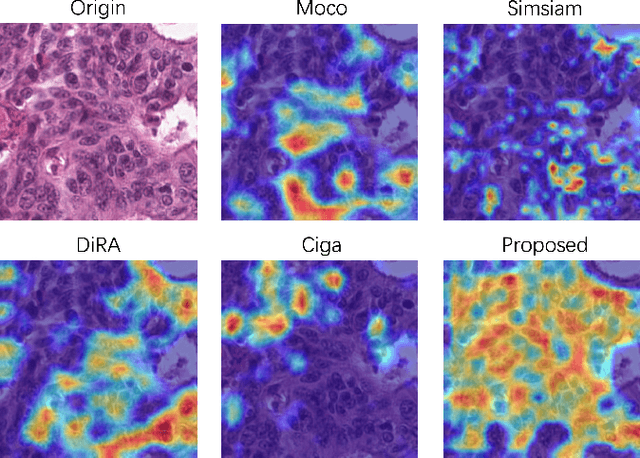
Abstract:Self-supervised pretraining attempts to enhance model performance by obtaining effective features from unlabeled data, and has demonstrated its effectiveness in the field of histopathology images. Despite its success, few works concentrate on the extraction of nucleus-level information, which is essential for pathologic analysis. In this work, we propose a novel nucleus-aware self-supervised pretraining framework for histopathology images. The framework aims to capture the nuclear morphology and distribution information through unpaired image-to-image translation between histopathology images and pseudo mask images. The generation process is modulated by both conditional and stochastic style representations, ensuring the reality and diversity of the generated histopathology images for pretraining. Further, an instance segmentation guided strategy is employed to capture instance-level information. The experiments on 7 datasets show that the proposed pretraining method outperforms supervised ones on Kather classification, multiple instance learning, and 5 dense-prediction tasks with the transfer learning protocol, and yields superior results than other self-supervised approaches on 8 semi-supervised tasks. Our project is publicly available at https://github.com/zhiyuns/UNITPathSSL.
COLosSAL: A Benchmark for Cold-start Active Learning for 3D Medical Image Segmentation
Jul 22, 2023Abstract:Medical image segmentation is a critical task in medical image analysis. In recent years, deep learning based approaches have shown exceptional performance when trained on a fully-annotated dataset. However, data annotation is often a significant bottleneck, especially for 3D medical images. Active learning (AL) is a promising solution for efficient annotation but requires an initial set of labeled samples to start active selection. When the entire data pool is unlabeled, how do we select the samples to annotate as our initial set? This is also known as the cold-start AL, which permits only one chance to request annotations from experts without access to previously annotated data. Cold-start AL is highly relevant in many practical scenarios but has been under-explored, especially for 3D medical segmentation tasks requiring substantial annotation effort. In this paper, we present a benchmark named COLosSAL by evaluating six cold-start AL strategies on five 3D medical image segmentation tasks from the public Medical Segmentation Decathlon collection. We perform a thorough performance analysis and explore important open questions for cold-start AL, such as the impact of budget on different strategies. Our results show that cold-start AL is still an unsolved problem for 3D segmentation tasks but some important trends have been observed. The code repository, data partitions, and baseline results for the complete benchmark are publicly available at https://github.com/MedICL-VU/COLosSAL.
DRMC: A Generalist Model with Dynamic Routing for Multi-Center PET Image Synthesis
Jul 11, 2023Abstract:Multi-center positron emission tomography (PET) image synthesis aims at recovering low-dose PET images from multiple different centers. The generalizability of existing methods can still be suboptimal for a multi-center study due to domain shifts, which result from non-identical data distribution among centers with different imaging systems/protocols. While some approaches address domain shifts by training specialized models for each center, they are parameter inefficient and do not well exploit the shared knowledge across centers. To address this, we develop a generalist model that shares architecture and parameters across centers to utilize the shared knowledge. However, the generalist model can suffer from the center interference issue, \textit{i.e.} the gradient directions of different centers can be inconsistent or even opposite owing to the non-identical data distribution. To mitigate such interference, we introduce a novel dynamic routing strategy with cross-layer connections that routes data from different centers to different experts. Experiments show that our generalist model with dynamic routing (DRMC) exhibits excellent generalizability across centers. Code and data are available at: https://github.com/Yaziwel/Multi-Center-PET-Image-Synthesis.
Zero-shot Nuclei Detection via Visual-Language Pre-trained Models
Jun 30, 2023



Abstract:Large-scale visual-language pre-trained models (VLPM) have proven their excellent performance in downstream object detection for natural scenes. However, zero-shot nuclei detection on H\&E images via VLPMs remains underexplored. The large gap between medical images and the web-originated text-image pairs used for pre-training makes it a challenging task. In this paper, we attempt to explore the potential of the object-level VLPM, Grounded Language-Image Pre-training (GLIP) model, for zero-shot nuclei detection. Concretely, an automatic prompts design pipeline is devised based on the association binding trait of VLPM and the image-to-text VLPM BLIP, avoiding empirical manual prompts engineering. We further establish a self-training framework, using the automatically designed prompts to generate the preliminary results as pseudo labels from GLIP and refine the predicted boxes in an iterative manner. Our method achieves a remarkable performance for label-free nuclei detection, surpassing other comparison methods. Foremost, our work demonstrates that the VLPM pre-trained on natural image-text pairs exhibits astonishing potential for downstream tasks in the medical field as well. Code will be released at https://github.com/wuyongjianCODE/VLPMNuD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge