Maode Lai
AttriPrompter: Auto-Prompting with Attribute Semantics for Zero-shot Nuclei Detection via Visual-Language Pre-trained Models
Oct 22, 2024



Abstract:Large-scale visual-language pre-trained models (VLPMs) have demonstrated exceptional performance in downstream object detection through text prompts for natural scenes. However, their application to zero-shot nuclei detection on histopathology images remains relatively unexplored, mainly due to the significant gap between the characteristics of medical images and the web-originated text-image pairs used for pre-training. This paper aims to investigate the potential of the object-level VLPM, Grounded Language-Image Pre-training (GLIP), for zero-shot nuclei detection. Specifically, we propose an innovative auto-prompting pipeline, named AttriPrompter, comprising attribute generation, attribute augmentation, and relevance sorting, to avoid subjective manual prompt design. AttriPrompter utilizes VLPMs' text-to-image alignment to create semantically rich text prompts, which are then fed into GLIP for initial zero-shot nuclei detection. Additionally, we propose a self-trained knowledge distillation framework, where GLIP serves as the teacher with its initial predictions used as pseudo labels, to address the challenges posed by high nuclei density, including missed detections, false positives, and overlapping instances. Our method exhibits remarkable performance in label-free nuclei detection, outperforming all existing unsupervised methods and demonstrating excellent generality. Notably, this work highlights the astonishing potential of VLPMs pre-trained on natural image-text pairs for downstream tasks in the medical field as well. Code will be released at https://github.com/wuyongjianCODE/AttriPrompter.
SDPT: Synchronous Dual Prompt Tuning for Fusion-based Visual-Language Pre-trained Models
Jul 16, 2024
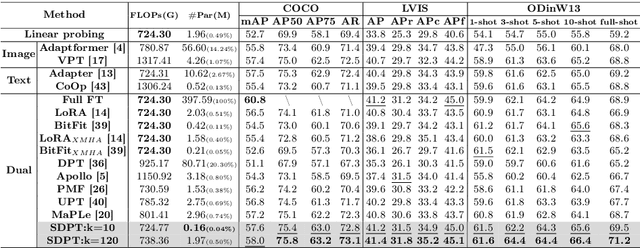
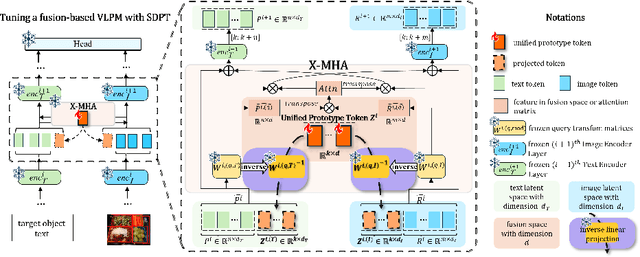

Abstract:Prompt tuning methods have achieved remarkable success in parameter-efficient fine-tuning on large pre-trained models. However, their application to dual-modal fusion-based visual-language pre-trained models (VLPMs), such as GLIP, has encountered issues. Existing prompt tuning methods have not effectively addressed the modal mapping and aligning problem for tokens in different modalities, leading to poor transfer generalization. To address this issue, we propose Synchronous Dual Prompt Tuning (SDPT). SDPT initializes a single set of learnable unified prototype tokens in the established modal aligning space to represent the aligned semantics of text and image modalities for downstream tasks. Furthermore, SDPT establishes inverse linear projections that require no training to embed the information of unified prototype tokens into the input space of different modalities. The inverse linear projections allow the unified prototype token to synchronously represent the two modalities and enable SDPT to share the unified semantics of text and image for downstream tasks across different modal prompts. Experimental results demonstrate that SDPT assists fusion-based VLPMs to achieve superior outcomes with only 0.04\% of model parameters for training across various scenarios, outperforming other single- or dual-modal methods. The code will be released at https://github.com/wuyongjianCODE/SDPT.
RCdpia: A Renal Carcinoma Digital Pathology Image Annotation dataset based on pathologists
Mar 17, 2024



Abstract:The annotation of digital pathological slide data for renal cell carcinoma is of paramount importance for correct diagnosis of artificial intelligence models due to the heterogeneous nature of the tumor. This process not only facilitates a deeper understanding of renal cell cancer heterogeneity but also aims to minimize noise in the data for more accurate studies. To enhance the applicability of the data, two pathologists were enlisted to meticulously curate, screen, and label a kidney cancer pathology image dataset from The Cancer Genome Atlas Program (TCGA) database. Subsequently, a Resnet model was developed to validate the annotated dataset against an additional dataset from the First Affiliated Hospital of Zhejiang University. Based on these results, we have meticulously compiled the TCGA digital pathological dataset with independent labeling of tumor regions and adjacent areas (RCdpia), which includes 109 cases of kidney chromophobe cell carcinoma, 486 cases of kidney clear cell carcinoma, and 292 cases of kidney papillary cell carcinoma. This dataset is now publicly accessible at http://39.171.241.18:8888/RCdpia/. Furthermore, model analysis has revealed significant discrepancies in predictive outcomes when applying the same model to datasets from different centers. Leveraging the RCdpia, we can now develop more precise digital pathology artificial intelligence models for tasks such as normalization, classification, and segmentation. These advancements underscore the potential for more nuanced and accurate AI applications in the field of digital pathology.
Nucleus-aware Self-supervised Pretraining Using Unpaired Image-to-image Translation for Histopathology Images
Sep 14, 2023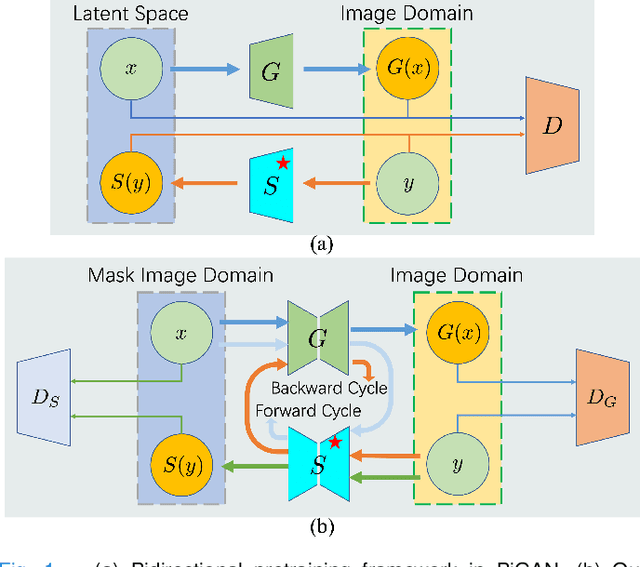
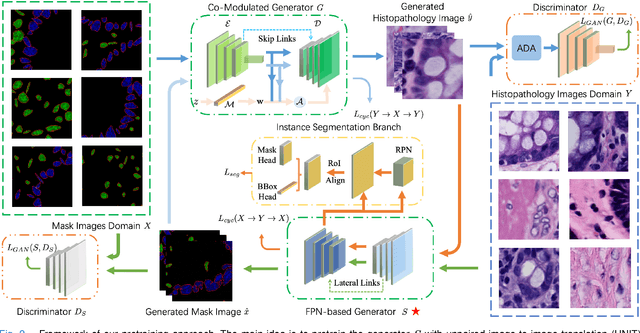
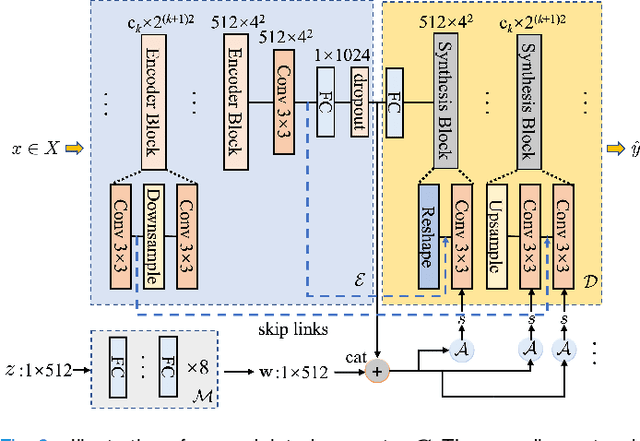
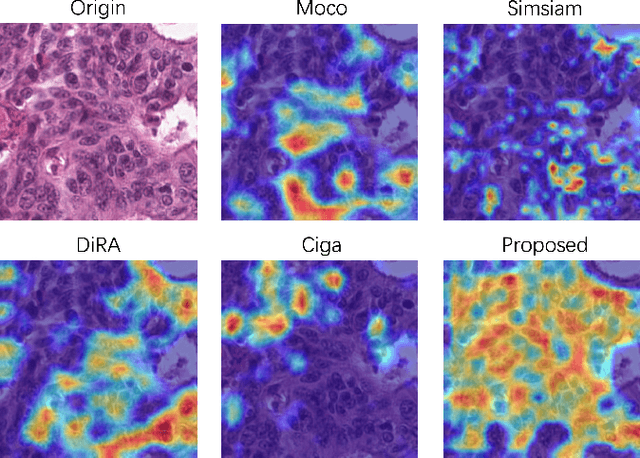
Abstract:Self-supervised pretraining attempts to enhance model performance by obtaining effective features from unlabeled data, and has demonstrated its effectiveness in the field of histopathology images. Despite its success, few works concentrate on the extraction of nucleus-level information, which is essential for pathologic analysis. In this work, we propose a novel nucleus-aware self-supervised pretraining framework for histopathology images. The framework aims to capture the nuclear morphology and distribution information through unpaired image-to-image translation between histopathology images and pseudo mask images. The generation process is modulated by both conditional and stochastic style representations, ensuring the reality and diversity of the generated histopathology images for pretraining. Further, an instance segmentation guided strategy is employed to capture instance-level information. The experiments on 7 datasets show that the proposed pretraining method outperforms supervised ones on Kather classification, multiple instance learning, and 5 dense-prediction tasks with the transfer learning protocol, and yields superior results than other self-supervised approaches on 8 semi-supervised tasks. Our project is publicly available at https://github.com/zhiyuns/UNITPathSSL.
Zero-shot Nuclei Detection via Visual-Language Pre-trained Models
Jun 30, 2023



Abstract:Large-scale visual-language pre-trained models (VLPM) have proven their excellent performance in downstream object detection for natural scenes. However, zero-shot nuclei detection on H\&E images via VLPMs remains underexplored. The large gap between medical images and the web-originated text-image pairs used for pre-training makes it a challenging task. In this paper, we attempt to explore the potential of the object-level VLPM, Grounded Language-Image Pre-training (GLIP) model, for zero-shot nuclei detection. Concretely, an automatic prompts design pipeline is devised based on the association binding trait of VLPM and the image-to-text VLPM BLIP, avoiding empirical manual prompts engineering. We further establish a self-training framework, using the automatically designed prompts to generate the preliminary results as pseudo labels from GLIP and refine the predicted boxes in an iterative manner. Our method achieves a remarkable performance for label-free nuclei detection, surpassing other comparison methods. Foremost, our work demonstrates that the VLPM pre-trained on natural image-text pairs exhibits astonishing potential for downstream tasks in the medical field as well. Code will be released at https://github.com/wuyongjianCODE/VLPMNuD.
Cyclic Learning: Bridging Image-level Labels and Nuclei Instance Segmentation
Jun 05, 2023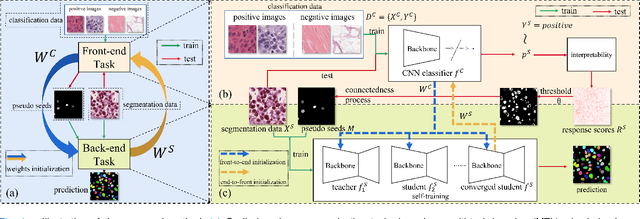
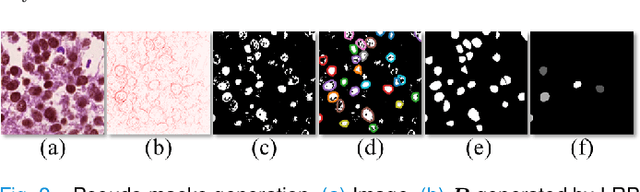
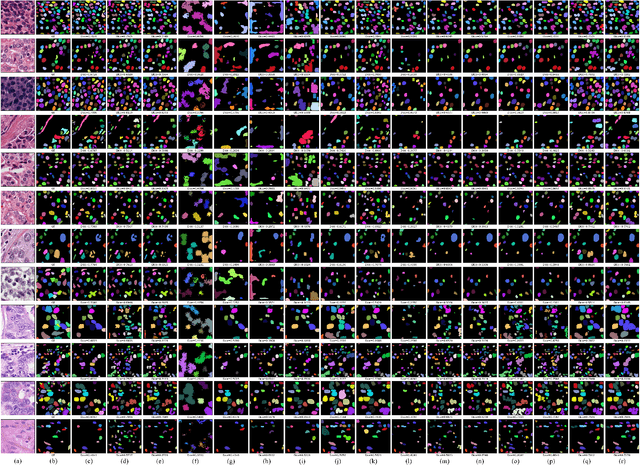
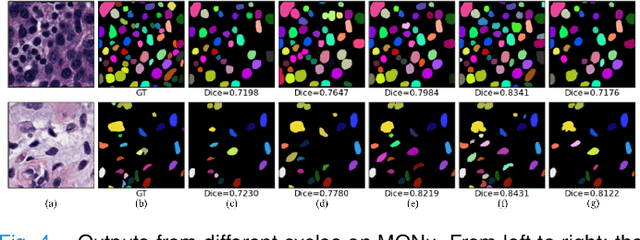
Abstract:Nuclei instance segmentation on histopathology images is of great clinical value for disease analysis. Generally, fully-supervised algorithms for this task require pixel-wise manual annotations, which is especially time-consuming and laborious for the high nuclei density. To alleviate the annotation burden, we seek to solve the problem through image-level weakly supervised learning, which is underexplored for nuclei instance segmentation. Compared with most existing methods using other weak annotations (scribble, point, etc.) for nuclei instance segmentation, our method is more labor-saving. The obstacle to using image-level annotations in nuclei instance segmentation is the lack of adequate location information, leading to severe nuclei omission or overlaps. In this paper, we propose a novel image-level weakly supervised method, called cyclic learning, to solve this problem. Cyclic learning comprises a front-end classification task and a back-end semi-supervised instance segmentation task to benefit from multi-task learning (MTL). We utilize a deep learning classifier with interpretability as the front-end to convert image-level labels to sets of high-confidence pseudo masks and establish a semi-supervised architecture as the back-end to conduct nuclei instance segmentation under the supervision of these pseudo masks. Most importantly, cyclic learning is designed to circularly share knowledge between the front-end classifier and the back-end semi-supervised part, which allows the whole system to fully extract the underlying information from image-level labels and converge to a better optimum. Experiments on three datasets demonstrate the good generality of our method, which outperforms other image-level weakly supervised methods for nuclei instance segmentation, and achieves comparable performance to fully-supervised methods.
Transformer based multiple instance learning for weakly supervised histopathology image segmentation
May 18, 2022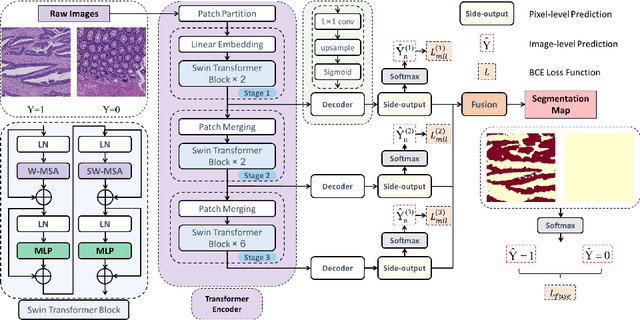
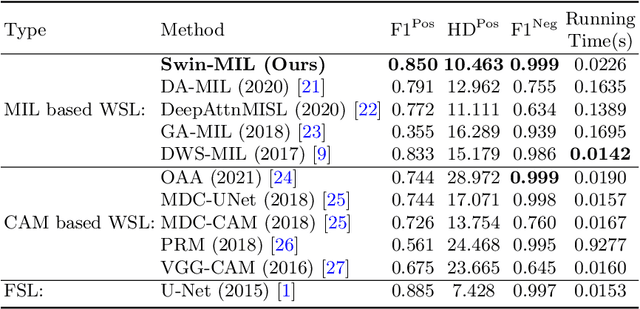
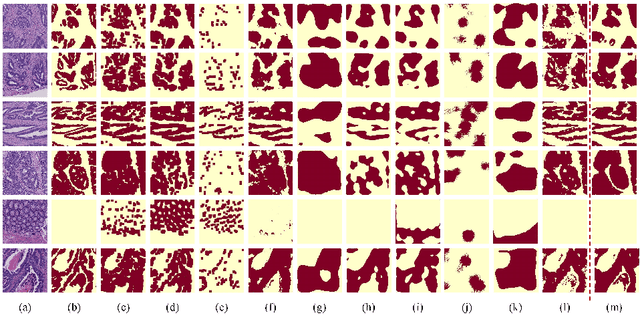
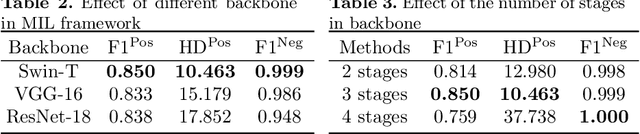
Abstract:Hispathological image segmentation algorithms play a critical role in computer aided diagnosis technology. The development of weakly supervised segmentation algorithm alleviates the problem of medical image annotation that it is time-consuming and labor-intensive. As a subset of weakly supervised learning, Multiple Instance Learning (MIL) has been proven to be effective in segmentation. However, there is a lack of related information between instances in MIL, which limits the further improvement of segmentation performance. In this paper, we propose a novel weakly supervised method for pixel-level segmentation in histopathology images, which introduces Transformer into the MIL framework to capture global or long-range dependencies. The multi-head self-attention in the Transformer establishes the relationship between instances, which solves the shortcoming that instances are independent of each other in MIL. In addition, deep supervision is introduced to overcome the limitation of annotations in weakly supervised methods and make the better utilization of hierarchical information. The state-of-the-art results on the colon cancer dataset demonstrate the superiority of the proposed method compared with other weakly supervised methods. It is worth believing that there is a potential of our approach for various applications in medical images.
Unsupervised Learning for Cell-level Visual Representation in Histopathology Images with Generative Adversarial Networks
Jul 07, 2018



Abstract:The visual attributes of cells, such as the nuclear morphology and chromatin openness, are critical for histopathology image analysis. By learning cell-level visual representation, we can obtain a rich mix of features that are highly reusable for various tasks, such as cell-level classification, nuclei segmentation, and cell counting. In this paper, we propose a unified generative adversarial networks architecture with a new formulation of loss to perform robust cell-level visual representation learning in an unsupervised setting. Our model is not only label-free and easily trained but also capable of cell-level unsupervised classification with interpretable visualization, which achieves promising results in the unsupervised classification of bone marrow cellular components. Based on the proposed cell-level visual representation learning, we further develop a pipeline that exploits the varieties of cellular elements to perform histopathology image classification, the advantages of which are demonstrated on bone marrow datasets.
Gland Instance Segmentation Using Deep Multichannel Neural Networks
Nov 23, 2017



Abstract:Objective: A new image instance segmentation method is proposed to segment individual glands (instances) in colon histology images. This process is challenging since the glands not only need to be segmented from a complex background, they must also be individually identified. Methods: We leverage the idea of image-to-image prediction in recent deep learning by designing an algorithm that automatically exploits and fuses complex multichannel information - regional, location, and boundary cues - in gland histology images. Our proposed algorithm, a deep multichannel framework, alleviates heavy feature design due to the use of convolutional neural networks and is able to meet multifarious requirements by altering channels. Results: Compared with methods reported in the 2015 MICCAI Gland Segmentation Challenge and other currently prevalent instance segmentation methods, we observe state-of-the-art results based on the evaluation metrics. Conclusion: The proposed deep multichannel algorithm is an effective method for gland instance segmentation. Significance: The generalization ability of our model not only enable the algorithm to solve gland instance segmentation problems, but the channel is also alternative that can be replaced for a specific task.
* arXiv admin note: substantial text overlap with arXiv:1607.04889
Gland Instance Segmentation by Deep Multichannel Neural Networks
Jul 19, 2016



Abstract:In this paper, we propose a new image instance segmentation method that segments individual glands (instances) in colon histology images. This is a task called instance segmentation that has recently become increasingly important. The problem is challenging since not only do the glands need to be segmented from the complex background, they are also required to be individually identified. Here we leverage the idea of image-to-image prediction in recent deep learning by building a framework that automatically exploits and fuses complex multichannel information, regional, location and boundary patterns in gland histology images. Our proposed system, deep multichannel framework, alleviates heavy feature design due to the use of convolutional neural networks and is able to meet multifarious requirement by altering channels. Compared to methods reported in the 2015 MICCAI Gland Segmentation Challenge and other currently prevalent methods of instance segmentation, we observe state-of-the-art results based on a number of evaluation metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge