Ziniu Qian
CTIS-QA: Clinical Template-Informed Slide-level Question Answering for Pathology
Jan 05, 2026Abstract:In this paper, we introduce a clinical diagnosis template-based pipeline to systematically collect and structure pathological information. In collaboration with pathologists and guided by the the College of American Pathologists (CAP) Cancer Protocols, we design a Clinical Pathology Report Template (CPRT) that ensures comprehensive and standardized extraction of diagnostic elements from pathology reports. We validate the effectiveness of our pipeline on TCGA-BRCA. First, we extract pathological features from reports using CPRT. These features are then used to build CTIS-Align, a dataset of 80k slide-description pairs from 804 WSIs for vision-language alignment training, and CTIS-Bench, a rigorously curated VQA benchmark comprising 977 WSIs and 14,879 question-answer pairs. CTIS-Bench emphasizes clinically grounded, closed-ended questions (e.g., tumor grade, receptor status) that reflect real diagnostic workflows, minimize non-visual reasoning, and require genuine slide understanding. We further propose CTIS-QA, a Slide-level Question Answering model, featuring a dual-stream architecture that mimics pathologists' diagnostic approach. One stream captures global slide-level context via clustering-based feature aggregation, while the other focuses on salient local regions through attention-guided patch perception module. Extensive experiments on WSI-VQA, CTIS-Bench, and slide-level diagnostic tasks show that CTIS-QA consistently outperforms existing state-of-the-art models across multiple metrics. Code and data are available at https://github.com/HLSvois/CTIS-QA.
Region Attention Transformer for Medical Image Restoration
Jul 12, 2024



Abstract:Transformer-based methods have demonstrated impressive results in medical image restoration, attributed to the multi-head self-attention (MSA) mechanism in the spatial dimension. However, the majority of existing Transformers conduct attention within fixed and coarsely partitioned regions (\text{e.g.} the entire image or fixed patches), resulting in interference from irrelevant regions and fragmentation of continuous image content. To overcome these challenges, we introduce a novel Region Attention Transformer (RAT) that utilizes a region-based multi-head self-attention mechanism (R-MSA). The R-MSA dynamically partitions the input image into non-overlapping semantic regions using the robust Segment Anything Model (SAM) and then performs self-attention within these regions. This region partitioning is more flexible and interpretable, ensuring that only pixels from similar semantic regions complement each other, thereby eliminating interference from irrelevant regions. Moreover, we introduce a focal region loss to guide our model to adaptively focus on recovering high-difficulty regions. Extensive experiments demonstrate the effectiveness of RAT in various medical image restoration tasks, including PET image synthesis, CT image denoising, and pathological image super-resolution. Code is available at \href{https://github.com/Yaziwel/Region-Attention-Transformer-for-Medical-Image-Restoration.git}{https://github.com/RAT}.
All-In-One Medical Image Restoration via Task-Adaptive Routing
May 30, 2024



Abstract:Although single-task medical image restoration (MedIR) has witnessed remarkable success, the limited generalizability of these methods poses a substantial obstacle to wider application. In this paper, we focus on the task of all-in-one medical image restoration, aiming to address multiple distinct MedIR tasks with a single universal model. Nonetheless, due to significant differences between different MedIR tasks, training a universal model often encounters task interference issues, where different tasks with shared parameters may conflict with each other in the gradient update direction. This task interference leads to deviation of the model update direction from the optimal path, thereby affecting the model's performance. To tackle this issue, we propose a task-adaptive routing strategy, allowing conflicting tasks to select different network paths in spatial and channel dimensions, thereby mitigating task interference. Experimental results demonstrate that our proposed \textbf{A}ll-in-one \textbf{M}edical \textbf{I}mage \textbf{R}estoration (\textbf{AMIR}) network achieves state-of-the-art performance in three MedIR tasks: MRI super-resolution, CT denoising, and PET synthesis, both in single-task and all-in-one settings. The code and data will be available at \href{https://github.com/Yaziwel/All-In-One-Medical-Image-Restoration-via-Task-Adaptive-Routing.git}{https://github.com/Yaziwel/AMIR}.
Transformer based multiple instance learning for weakly supervised histopathology image segmentation
May 18, 2022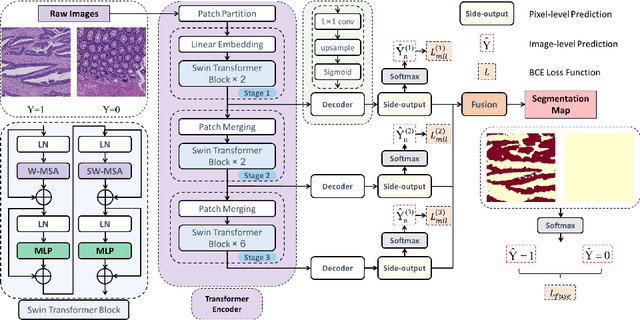
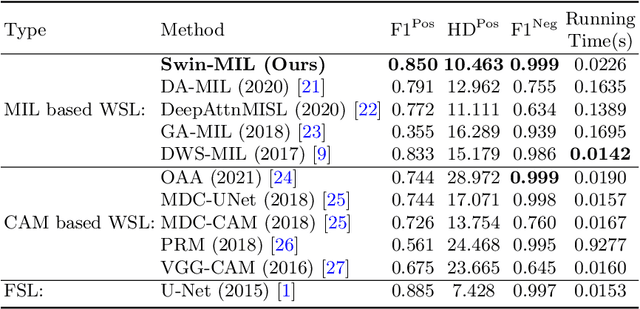
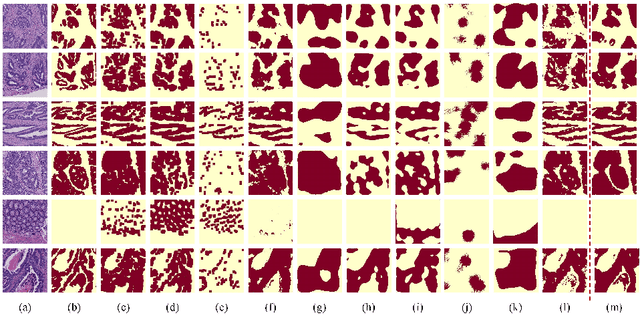
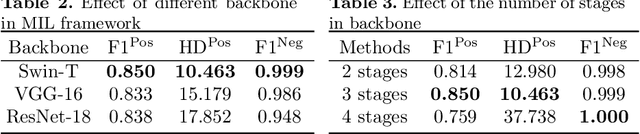
Abstract:Hispathological image segmentation algorithms play a critical role in computer aided diagnosis technology. The development of weakly supervised segmentation algorithm alleviates the problem of medical image annotation that it is time-consuming and labor-intensive. As a subset of weakly supervised learning, Multiple Instance Learning (MIL) has been proven to be effective in segmentation. However, there is a lack of related information between instances in MIL, which limits the further improvement of segmentation performance. In this paper, we propose a novel weakly supervised method for pixel-level segmentation in histopathology images, which introduces Transformer into the MIL framework to capture global or long-range dependencies. The multi-head self-attention in the Transformer establishes the relationship between instances, which solves the shortcoming that instances are independent of each other in MIL. In addition, deep supervision is introduced to overcome the limitation of annotations in weakly supervised methods and make the better utilization of hierarchical information. The state-of-the-art results on the colon cancer dataset demonstrate the superiority of the proposed method compared with other weakly supervised methods. It is worth believing that there is a potential of our approach for various applications in medical images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge